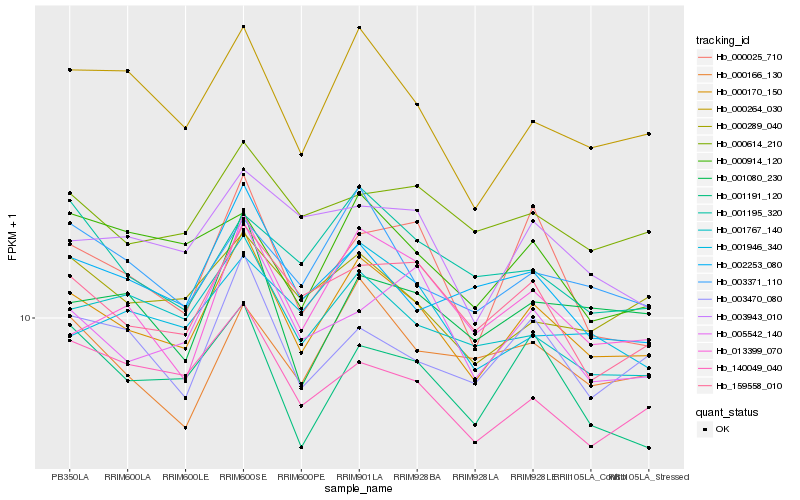
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000170_150 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638996 isoform X3 [Jatropha curcas] |
| 2 |
Hb_000264_030 |
0.0546828644 |
- |
- |
PREDICTED: splicing factor 3B subunit 2 [Jatropha curcas] |
| 3 |
Hb_159558_010 |
0.0597227486 |
- |
- |
PREDICTED: uncharacterized protein LOC105635846 [Jatropha curcas] |
| 4 |
Hb_001767_140 |
0.0612058792 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |
| 5 |
Hb_003371_110 |
0.061958989 |
- |
- |
PREDICTED: protein EMSY-LIKE 3 [Jatropha curcas] |
| 6 |
Hb_003470_080 |
0.0632784213 |
transcription factor |
TF Family: SNF2 |
PREDICTED: protein PHOTOPERIOD-INDEPENDENT EARLY FLOWERING 1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000289_040 |
0.065256918 |
- |
- |
Poly(A) polymerase alpha, putative [Ricinus communis] |
| 8 |
Hb_002253_080 |
0.0653970622 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase beta 1-like [Jatropha curcas] |
| 9 |
Hb_000614_210 |
0.0657669328 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-1-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_140049_040 |
0.0663767366 |
- |
- |
PREDICTED: uncharacterized protein LOC105632012 [Jatropha curcas] |
| 11 |
Hb_000025_710 |
0.0664846252 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |
| 12 |
Hb_001191_120 |
0.0666987407 |
- |
- |
PREDICTED: cleavage stimulating factor 64 isoform X1 [Jatropha curcas] |
| 13 |
Hb_013399_070 |
0.0670067586 |
- |
- |
PREDICTED: uncharacterized protein LOC105631409 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003943_010 |
0.0678021978 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_005542_140 |
0.0683619688 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 16 |
Hb_001080_230 |
0.0684592358 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105649979 [Jatropha curcas] |
| 17 |
Hb_000914_120 |
0.068950426 |
- |
- |
PREDICTED: outer envelope protein 80, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000166_130 |
0.0701787191 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001946_340 |
0.070417263 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 17-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_001195_320 |
0.0718507451 |
- |
- |
PREDICTED: S-adenosyl-L-methionine-dependent tRNA 4-demethylwyosine synthase [Jatropha curcas] |