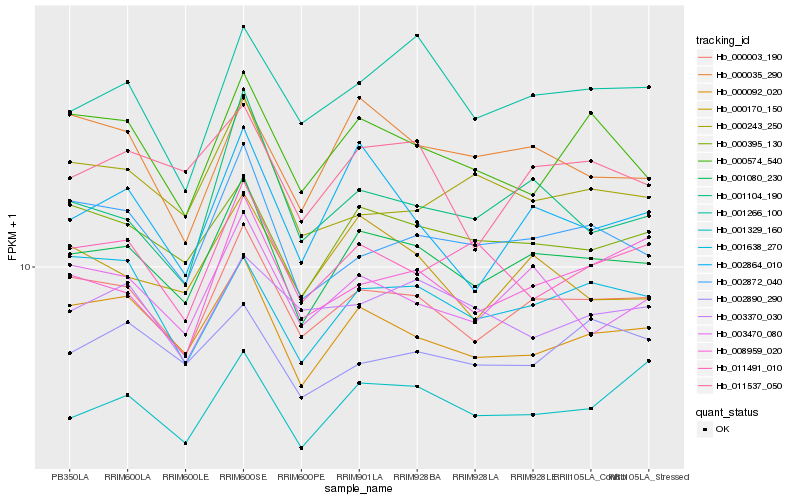
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000003_190 |
0.0 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA-like isoform X1 [Nelumbo nucifera] |
| 2 |
Hb_001080_230 |
0.0404081052 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105649979 [Jatropha curcas] |
| 3 |
Hb_000092_020 |
0.0521079939 |
transcription factor |
TF Family: MYB |
transcription factor, putative [Ricinus communis] |
| 4 |
Hb_003470_080 |
0.0535135706 |
transcription factor |
TF Family: SNF2 |
PREDICTED: protein PHOTOPERIOD-INDEPENDENT EARLY FLOWERING 1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002872_040 |
0.0570300973 |
- |
- |
pumilio, putative [Ricinus communis] |
| 6 |
Hb_000395_130 |
0.0589744518 |
- |
- |
PREDICTED: N6-adenosine-methyltransferase MT-A70-like [Jatropha curcas] |
| 7 |
Hb_008959_020 |
0.0603056285 |
- |
- |
PREDICTED: nitric oxide synthase-interacting protein [Pyrus x bretschneideri] |
| 8 |
Hb_001638_270 |
0.0632539552 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 8 homolog isoform X2 [Jatropha curcas] |
| 9 |
Hb_000574_540 |
0.0674242051 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_000170_150 |
0.0731194344 |
- |
- |
PREDICTED: uncharacterized protein LOC105638996 isoform X3 [Jatropha curcas] |
| 11 |
Hb_011537_050 |
0.0731658495 |
- |
- |
PREDICTED: protein EMSY-LIKE 3 isoform X1 [Jatropha curcas] |
| 12 |
Hb_011491_010 |
0.07319896 |
- |
- |
Protein YME1, putative [Ricinus communis] |
| 13 |
Hb_001104_190 |
0.0736872983 |
- |
- |
PREDICTED: uncharacterized protein LOC105629451 [Jatropha curcas] |
| 14 |
Hb_001266_100 |
0.0745501929 |
- |
- |
PREDICTED: uncharacterized protein LOC105649131 [Jatropha curcas] |
| 15 |
Hb_000243_250 |
0.0748207281 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 44 [Jatropha curcas] |
| 16 |
Hb_003370_030 |
0.0760607114 |
- |
- |
PREDICTED: uncharacterized protein LOC105635547 isoform X2 [Jatropha curcas] |
| 17 |
Hb_001329_160 |
0.0770864775 |
- |
- |
PREDICTED: uncharacterized protein LOC105642679 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000035_290 |
0.0771211187 |
- |
- |
RNA recognition motif-containing family protein [Populus trichocarpa] |
| 19 |
Hb_002864_010 |
0.0781026808 |
- |
- |
PREDICTED: regulator of nonsense transcripts 1 homolog [Jatropha curcas] |
| 20 |
Hb_002890_290 |
0.07852254 |
- |
- |
PREDICTED: outer envelope protein 64, mitochondrial [Jatropha curcas] |