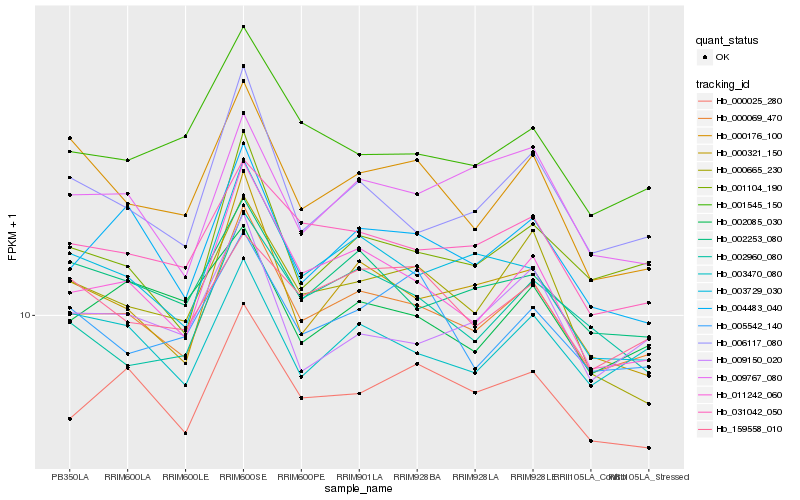
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000069_470 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 2 |
Hb_011242_060 |
0.0458698467 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 3 |
Hb_031042_050 |
0.0515165032 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 4 |
Hb_006117_080 |
0.0548372589 |
- |
- |
PREDICTED: transcription factor GTE4 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000025_280 |
0.0554872841 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_07432 [Jatropha curcas] |
| 6 |
Hb_001104_190 |
0.0557552351 |
- |
- |
PREDICTED: uncharacterized protein LOC105629451 [Jatropha curcas] |
| 7 |
Hb_004483_040 |
0.0557710214 |
- |
- |
hypothetical protein JCGZ_22047 [Jatropha curcas] |
| 8 |
Hb_002253_080 |
0.0581026876 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase beta 1-like [Jatropha curcas] |
| 9 |
Hb_000321_150 |
0.0591399107 |
- |
- |
PREDICTED: uncharacterized protein LOC105645596 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003729_030 |
0.0627740707 |
- |
- |
PREDICTED: alpha,alpha-trehalose-phosphate synthase [UDP-forming] 6 [Jatropha curcas] |
| 11 |
Hb_003470_080 |
0.0628246483 |
transcription factor |
TF Family: SNF2 |
PREDICTED: protein PHOTOPERIOD-INDEPENDENT EARLY FLOWERING 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001545_150 |
0.0660166324 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105635527 [Jatropha curcas] |
| 13 |
Hb_159558_010 |
0.0667546233 |
- |
- |
PREDICTED: uncharacterized protein LOC105635846 [Jatropha curcas] |
| 14 |
Hb_000176_100 |
0.0671741019 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Solanum lycopersicum] |
| 15 |
Hb_009767_080 |
0.0682321321 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105631983 [Jatropha curcas] |
| 16 |
Hb_002960_080 |
0.0689077269 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105635322 [Jatropha curcas] |
| 17 |
Hb_000665_230 |
0.0696385381 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 18 |
Hb_002085_030 |
0.0706325526 |
- |
- |
PREDICTED: protein CHROMATIN REMODELING 4 isoform X2 [Jatropha curcas] |
| 19 |
Hb_009150_020 |
0.0707676401 |
- |
- |
PREDICTED: uncharacterized protein LOC105630583 [Jatropha curcas] |
| 20 |
Hb_005542_140 |
0.0708595951 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |