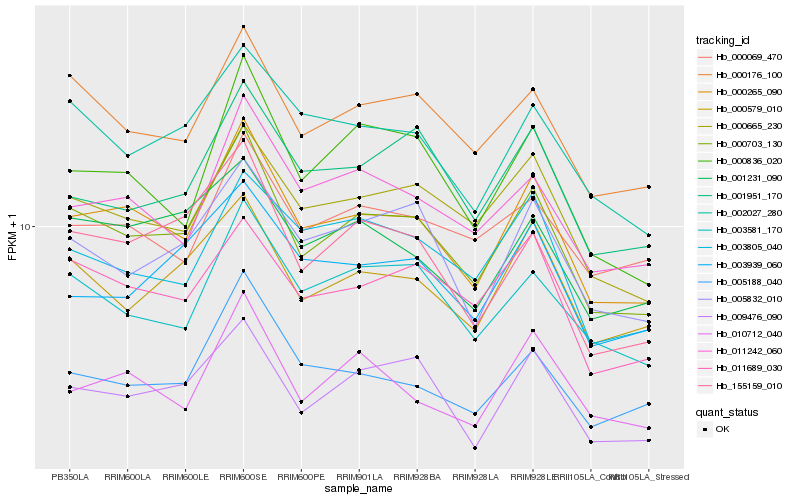
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000265_090 |
0.0 |
- |
- |
PREDICTED: flowering time control protein FPA [Jatropha curcas] |
| 2 |
Hb_000703_130 |
0.0434623051 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 [Jatropha curcas] |
| 3 |
Hb_011242_060 |
0.064080928 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 4 |
Hb_000665_230 |
0.0685402992 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 5 |
Hb_005188_040 |
0.0696334215 |
- |
- |
PREDICTED: protein furry homolog-like [Nicotiana sylvestris] |
| 6 |
Hb_010712_040 |
0.0722669536 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g11290 [Jatropha curcas] |
| 7 |
Hb_000836_020 |
0.0724437043 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 8 |
Hb_009476_090 |
0.0739363569 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 1 [Jatropha curcas] |
| 9 |
Hb_000176_100 |
0.0744913581 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Solanum lycopersicum] |
| 10 |
Hb_003581_170 |
0.0751161612 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_002027_280 |
0.0756360576 |
- |
- |
PREDICTED: sister-chromatid cohesion protein 3 [Jatropha curcas] |
| 12 |
Hb_011689_030 |
0.0758078611 |
- |
- |
80 kD MCM3-associated protein, putative [Ricinus communis] |
| 13 |
Hb_001231_090 |
0.0759060747 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 14 |
Hb_005832_010 |
0.0782119594 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 15 |
Hb_003805_040 |
0.079747375 |
- |
- |
Sacsin [Glycine soja] |
| 16 |
Hb_000069_470 |
0.0800923291 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000579_010 |
0.0807882656 |
transcription factor |
TF Family: SET |
PREDICTED: probable histone-lysine N-methyltransferase ATXR3 [Jatropha curcas] |
| 18 |
Hb_155159_010 |
0.0819783141 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 1 [Jatropha curcas] |
| 19 |
Hb_001951_170 |
0.0822220485 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 20 |
Hb_003939_060 |
0.0829036084 |
- |
- |
conserved hypothetical protein [Ricinus communis] |