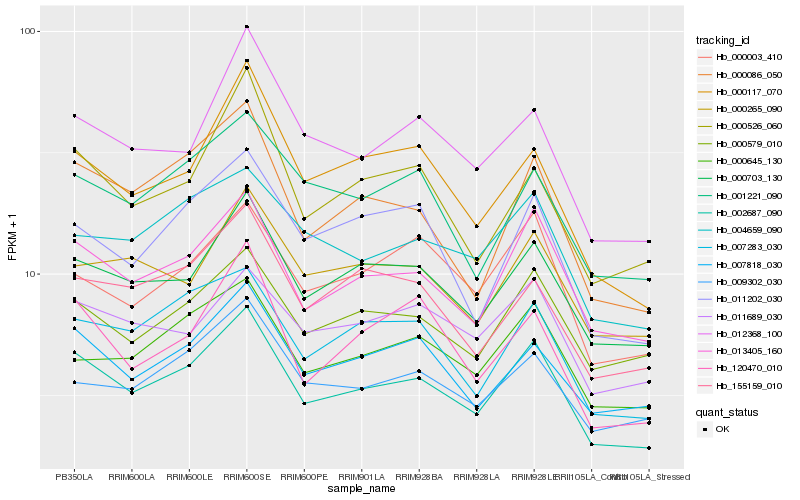
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002687_090 |
0.0 |
- |
- |
PREDICTED: PAX3- and PAX7-binding protein 1 [Jatropha curcas] |
| 2 |
Hb_013405_160 |
0.0483685933 |
- |
- |
PREDICTED: methyltransferase-like protein 1 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000703_130 |
0.0639246455 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 [Jatropha curcas] |
| 4 |
Hb_007818_030 |
0.0745945919 |
- |
- |
PREDICTED: transcriptional corepressor SEUSS-like [Jatropha curcas] |
| 5 |
Hb_000086_050 |
0.0749815837 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 1 [Jatropha curcas] |
| 6 |
Hb_000579_010 |
0.0761625759 |
transcription factor |
TF Family: SET |
PREDICTED: probable histone-lysine N-methyltransferase ATXR3 [Jatropha curcas] |
| 7 |
Hb_007283_030 |
0.0786204813 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40C-like [Malus domestica] |
| 8 |
Hb_000645_130 |
0.0816804975 |
- |
- |
PREDICTED: bromodomain and WD repeat-containing protein 3 [Jatropha curcas] |
| 9 |
Hb_009302_030 |
0.0847689874 |
- |
- |
hypothetical protein RCOM_1176340 [Ricinus communis] |
| 10 |
Hb_004659_090 |
0.0856431996 |
- |
- |
PREDICTED: uncharacterized protein LOC105629935 [Jatropha curcas] |
| 11 |
Hb_012368_100 |
0.0861516454 |
- |
- |
PREDICTED: uncharacterized protein LOC105637426 [Jatropha curcas] |
| 12 |
Hb_155159_010 |
0.0867678257 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 1 [Jatropha curcas] |
| 13 |
Hb_000117_070 |
0.0869002536 |
- |
- |
PREDICTED: branchpoint-bridging protein [Jatropha curcas] |
| 14 |
Hb_001221_090 |
0.0870738874 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |
| 15 |
Hb_011202_030 |
0.0878942866 |
- |
- |
arsenite-resistance protein, putative [Ricinus communis] |
| 16 |
Hb_011689_030 |
0.0880452581 |
- |
- |
80 kD MCM3-associated protein, putative [Ricinus communis] |
| 17 |
Hb_000003_410 |
0.0881924793 |
- |
- |
PREDICTED: intron-binding protein aquarius [Jatropha curcas] |
| 18 |
Hb_000526_060 |
0.0885690115 |
- |
- |
PREDICTED: clustered mitochondria protein [Jatropha curcas] |
| 19 |
Hb_000265_090 |
0.0890687239 |
- |
- |
PREDICTED: flowering time control protein FPA [Jatropha curcas] |
| 20 |
Hb_120470_010 |
0.090716649 |
- |
- |
PREDICTED: PAX3- and PAX7-binding protein 1 [Jatropha curcas] |