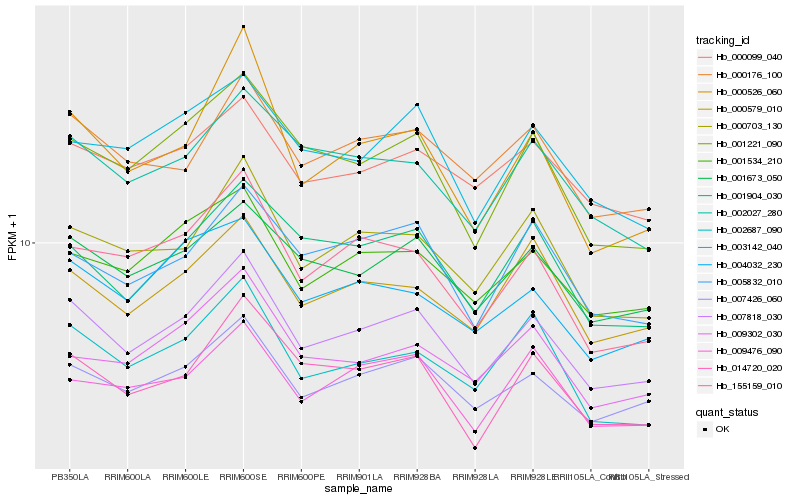
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007818_030 |
0.0 |
- |
- |
PREDICTED: transcriptional corepressor SEUSS-like [Jatropha curcas] |
| 2 |
Hb_007426_060 |
0.0502435002 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 3 |
Hb_000703_130 |
0.0600237235 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 [Jatropha curcas] |
| 4 |
Hb_001673_050 |
0.0633502656 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_001221_090 |
0.0637640636 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000526_060 |
0.0676817367 |
- |
- |
PREDICTED: clustered mitochondria protein [Jatropha curcas] |
| 7 |
Hb_000176_100 |
0.0693113278 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Solanum lycopersicum] |
| 8 |
Hb_001534_210 |
0.0694108843 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105642316 isoform X2 [Jatropha curcas] |
| 9 |
Hb_155159_010 |
0.0696804879 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 1 [Jatropha curcas] |
| 10 |
Hb_002027_280 |
0.0723026709 |
- |
- |
PREDICTED: sister-chromatid cohesion protein 3 [Jatropha curcas] |
| 11 |
Hb_001904_030 |
0.0723178493 |
- |
- |
PREDICTED: importin-5 [Jatropha curcas] |
| 12 |
Hb_014720_020 |
0.0739610993 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATM [Jatropha curcas] |
| 13 |
Hb_005832_010 |
0.0740846287 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000579_010 |
0.0741212622 |
transcription factor |
TF Family: SET |
PREDICTED: probable histone-lysine N-methyltransferase ATXR3 [Jatropha curcas] |
| 15 |
Hb_002687_090 |
0.0745945919 |
- |
- |
PREDICTED: PAX3- and PAX7-binding protein 1 [Jatropha curcas] |
| 16 |
Hb_004032_230 |
0.0771106194 |
- |
- |
PREDICTED: uncharacterized protein LOC105635143 [Jatropha curcas] |
| 17 |
Hb_009476_090 |
0.0771119239 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 1 [Jatropha curcas] |
| 18 |
Hb_003142_040 |
0.0785128596 |
- |
- |
Sf3a3 [Gossypium arboreum] |
| 19 |
Hb_009302_030 |
0.078827441 |
- |
- |
hypothetical protein RCOM_1176340 [Ricinus communis] |
| 20 |
Hb_000099_040 |
0.0804974003 |
- |
- |
unnamed protein product [Coffea canephora] |