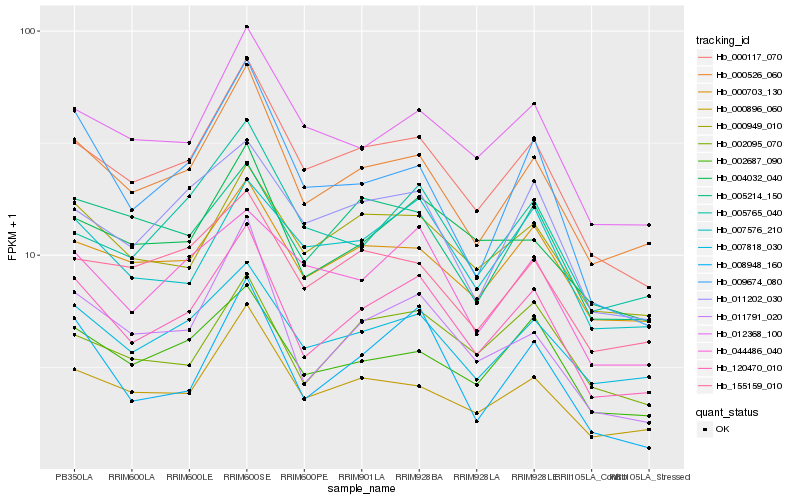
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_120470_010 |
0.0 |
- |
- |
PREDICTED: PAX3- and PAX7-binding protein 1 [Jatropha curcas] |
| 2 |
Hb_000526_060 |
0.0692040619 |
- |
- |
PREDICTED: clustered mitochondria protein [Jatropha curcas] |
| 3 |
Hb_000117_070 |
0.07514026 |
- |
- |
PREDICTED: branchpoint-bridging protein [Jatropha curcas] |
| 4 |
Hb_011791_020 |
0.0876238047 |
- |
- |
hypothetical protein Csa_6G486930 [Cucumis sativus] |
| 5 |
Hb_007818_030 |
0.0881254126 |
- |
- |
PREDICTED: transcriptional corepressor SEUSS-like [Jatropha curcas] |
| 6 |
Hb_002687_090 |
0.090716649 |
- |
- |
PREDICTED: PAX3- and PAX7-binding protein 1 [Jatropha curcas] |
| 7 |
Hb_004032_040 |
0.0933939128 |
- |
- |
unknown [Medicago truncatula] |
| 8 |
Hb_000896_060 |
0.0939737591 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_000949_010 |
0.0958305746 |
- |
- |
PREDICTED: regulator of nonsense transcripts UPF2 [Jatropha curcas] |
| 10 |
Hb_000703_130 |
0.099456579 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 [Jatropha curcas] |
| 11 |
Hb_008948_160 |
0.0994651852 |
- |
- |
PREDICTED: uncharacterized protein LOC104214688 isoform X2 [Nicotiana sylvestris] |
| 12 |
Hb_012368_100 |
0.0996894827 |
- |
- |
PREDICTED: uncharacterized protein LOC105637426 [Jatropha curcas] |
| 13 |
Hb_002095_070 |
0.1045679815 |
- |
- |
PREDICTED: probable pre-mRNA-splicing factor ATP-dependent RNA helicase [Jatropha curcas] |
| 14 |
Hb_011202_030 |
0.104781008 |
- |
- |
arsenite-resistance protein, putative [Ricinus communis] |
| 15 |
Hb_007576_210 |
0.1056625128 |
- |
- |
PREDICTED: protein furry homolog [Jatropha curcas] |
| 16 |
Hb_044486_040 |
0.1069249462 |
- |
- |
PREDICTED: ion channel CASTOR-like [Malus domestica] |
| 17 |
Hb_005214_150 |
0.1097870654 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105636019 [Jatropha curcas] |
| 18 |
Hb_009674_080 |
0.1099168674 |
- |
- |
PREDICTED: branchpoint-bridging protein [Jatropha curcas] |
| 19 |
Hb_155159_010 |
0.1100682806 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 1 [Jatropha curcas] |
| 20 |
Hb_005765_040 |
0.1104088769 |
- |
- |
transcription elongation factor s-II, putative [Ricinus communis] |