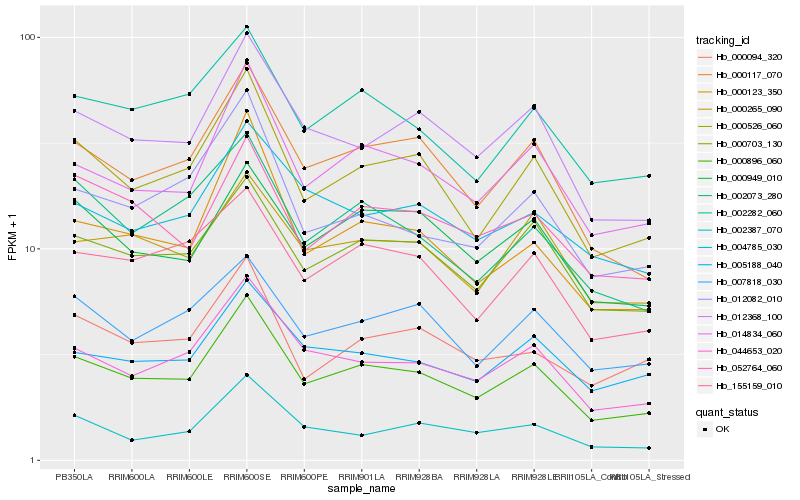
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000896_060 |
0.0 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_000526_060 |
0.0488666483 |
- |
- |
PREDICTED: clustered mitochondria protein [Jatropha curcas] |
| 3 |
Hb_014834_060 |
0.0561991903 |
- |
- |
PREDICTED: uncharacterized protein LOC105632277 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000117_070 |
0.0597894729 |
- |
- |
PREDICTED: branchpoint-bridging protein [Jatropha curcas] |
| 5 |
Hb_044653_020 |
0.0639027866 |
transcription factor, desease resistance |
TF Family: GNAT, Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 6 |
Hb_012368_100 |
0.0661635666 |
- |
- |
PREDICTED: uncharacterized protein LOC105637426 [Jatropha curcas] |
| 7 |
Hb_000123_350 |
0.070080246 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 8 |
Hb_000703_130 |
0.0733247632 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 [Jatropha curcas] |
| 9 |
Hb_002282_060 |
0.0772401227 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |
| 10 |
Hb_155159_010 |
0.0809425994 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 1 [Jatropha curcas] |
| 11 |
Hb_002073_280 |
0.0852991336 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 3 isoform X1 [Jatropha curcas] |
| 12 |
Hb_005188_040 |
0.0864259685 |
- |
- |
PREDICTED: protein furry homolog-like [Nicotiana sylvestris] |
| 13 |
Hb_012082_010 |
0.0864626575 |
- |
- |
PREDICTED: transcription factor GTE4 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000949_010 |
0.0866861282 |
- |
- |
PREDICTED: regulator of nonsense transcripts UPF2 [Jatropha curcas] |
| 15 |
Hb_052764_060 |
0.0879187442 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 16 |
Hb_007818_030 |
0.0891660644 |
- |
- |
PREDICTED: transcriptional corepressor SEUSS-like [Jatropha curcas] |
| 17 |
Hb_000265_090 |
0.0899656352 |
- |
- |
PREDICTED: flowering time control protein FPA [Jatropha curcas] |
| 18 |
Hb_000094_320 |
0.0918058254 |
- |
- |
hypothetical protein JCGZ_04407 [Jatropha curcas] |
| 19 |
Hb_004785_030 |
0.0919153797 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor VIP1 [Jatropha curcas] |
| 20 |
Hb_002387_070 |
0.0928574602 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g77010, mitochondrial [Jatropha curcas] |