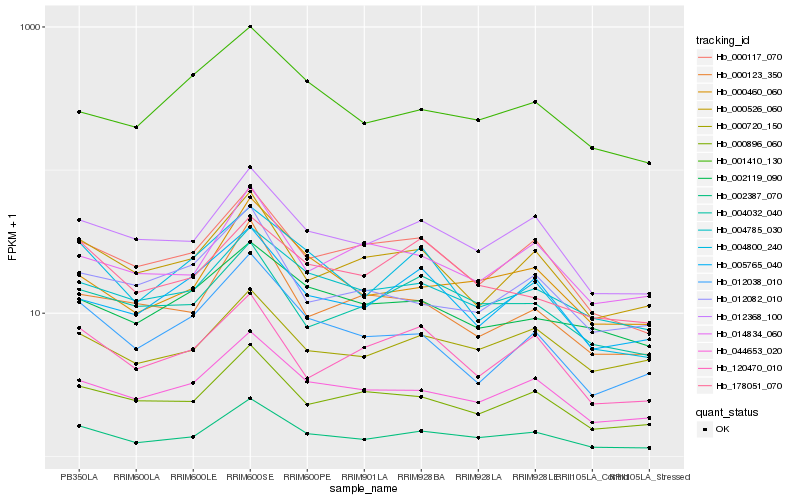
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002387_070 |
0.0 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g77010, mitochondrial [Jatropha curcas] |
| 2 |
Hb_012368_100 |
0.0747590493 |
- |
- |
PREDICTED: uncharacterized protein LOC105637426 [Jatropha curcas] |
| 3 |
Hb_044653_020 |
0.077493663 |
transcription factor, desease resistance |
TF Family: GNAT, Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 4 |
Hb_000460_060 |
0.0840891696 |
- |
- |
importin beta-1, putative [Ricinus communis] |
| 5 |
Hb_000896_060 |
0.0928574602 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_000117_070 |
0.0958580467 |
- |
- |
PREDICTED: branchpoint-bridging protein [Jatropha curcas] |
| 7 |
Hb_000526_060 |
0.1005227321 |
- |
- |
PREDICTED: clustered mitochondria protein [Jatropha curcas] |
| 8 |
Hb_004785_030 |
0.1024243332 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor VIP1 [Jatropha curcas] |
| 9 |
Hb_000720_150 |
0.1084059852 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X3 [Jatropha curcas] |
| 10 |
Hb_000123_350 |
0.1098291731 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 11 |
Hb_014834_060 |
0.1101788655 |
- |
- |
PREDICTED: uncharacterized protein LOC105632277 isoform X1 [Jatropha curcas] |
| 12 |
Hb_012038_010 |
0.1113987241 |
- |
- |
phospholipid-transporting atpase, putative [Ricinus communis] |
| 13 |
Hb_004032_040 |
0.113046434 |
- |
- |
unknown [Medicago truncatula] |
| 14 |
Hb_005765_040 |
0.1140661313 |
- |
- |
transcription elongation factor s-II, putative [Ricinus communis] |
| 15 |
Hb_120470_010 |
0.1162653998 |
- |
- |
PREDICTED: PAX3- and PAX7-binding protein 1 [Jatropha curcas] |
| 16 |
Hb_004800_240 |
0.120192954 |
- |
- |
PREDICTED: vam6/Vps39-like protein [Jatropha curcas] |
| 17 |
Hb_001410_130 |
0.1223831889 |
- |
- |
hypothetical protein POPTR_0001s10880g [Populus trichocarpa] |
| 18 |
Hb_002119_090 |
0.1224402714 |
- |
- |
PREDICTED: protein OBERON 3 [Jatropha curcas] |
| 19 |
Hb_012082_010 |
0.1228599716 |
- |
- |
PREDICTED: transcription factor GTE4 isoform X1 [Jatropha curcas] |
| 20 |
Hb_178051_070 |
0.1236853085 |
- |
- |
PREDICTED: exocyst complex component EXO70B1-like [Jatropha curcas] |