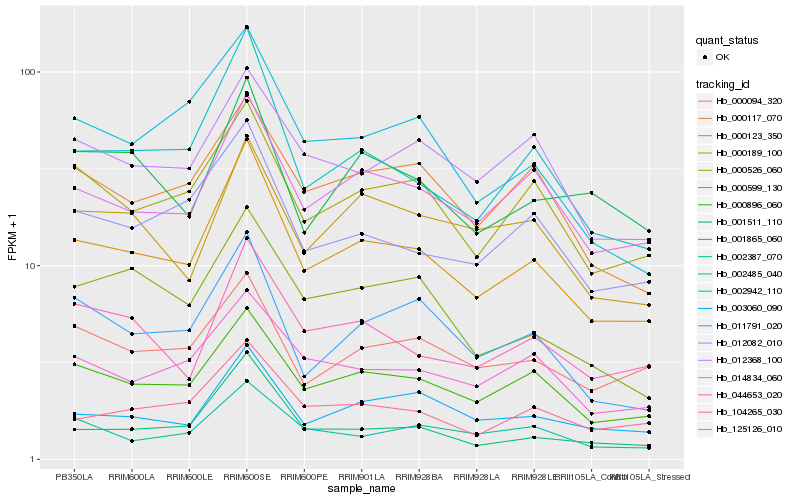
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000123_350 |
0.0 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 2 |
Hb_000896_060 |
0.070080246 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_002485_040 |
0.0739796455 |
- |
- |
PREDICTED: uncharacterized protein LOC105637426 [Jatropha curcas] |
| 4 |
Hb_014834_060 |
0.0839516364 |
- |
- |
PREDICTED: uncharacterized protein LOC105632277 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000526_060 |
0.0877644653 |
- |
- |
PREDICTED: clustered mitochondria protein [Jatropha curcas] |
| 6 |
Hb_003060_090 |
0.092831942 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 7 |
Hb_012082_010 |
0.0947083206 |
- |
- |
PREDICTED: transcription factor GTE4 isoform X1 [Jatropha curcas] |
| 8 |
Hb_125126_010 |
0.0990369602 |
desease resistance |
Gene Name: NB-ARC |
resistance family protein [Populus trichocarpa] |
| 9 |
Hb_000117_070 |
0.1009805937 |
- |
- |
PREDICTED: branchpoint-bridging protein [Jatropha curcas] |
| 10 |
Hb_001865_060 |
0.1012392807 |
- |
- |
PREDICTED: uncharacterized protein LOC105647962 [Jatropha curcas] |
| 11 |
Hb_044653_020 |
0.1018189613 |
transcription factor, desease resistance |
TF Family: GNAT, Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 12 |
Hb_011791_020 |
0.1023906849 |
- |
- |
hypothetical protein Csa_6G486930 [Cucumis sativus] |
| 13 |
Hb_104265_030 |
0.1038094349 |
- |
- |
hypothetical protein POPTR_0001s31230g [Populus trichocarpa] |
| 14 |
Hb_002942_110 |
0.10892223 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 2-like [Jatropha curcas] |
| 15 |
Hb_000599_130 |
0.1096506417 |
- |
- |
PREDICTED: U-box domain-containing protein 3 [Jatropha curcas] |
| 16 |
Hb_002387_070 |
0.1098291731 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g77010, mitochondrial [Jatropha curcas] |
| 17 |
Hb_012368_100 |
0.1106798814 |
- |
- |
PREDICTED: uncharacterized protein LOC105637426 [Jatropha curcas] |
| 18 |
Hb_000094_320 |
0.1113457144 |
- |
- |
hypothetical protein JCGZ_04407 [Jatropha curcas] |
| 19 |
Hb_000189_100 |
0.1144484769 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 14 [Jatropha curcas] |
| 20 |
Hb_001511_110 |
0.1159687404 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2 isoform X2 [Jatropha curcas] |