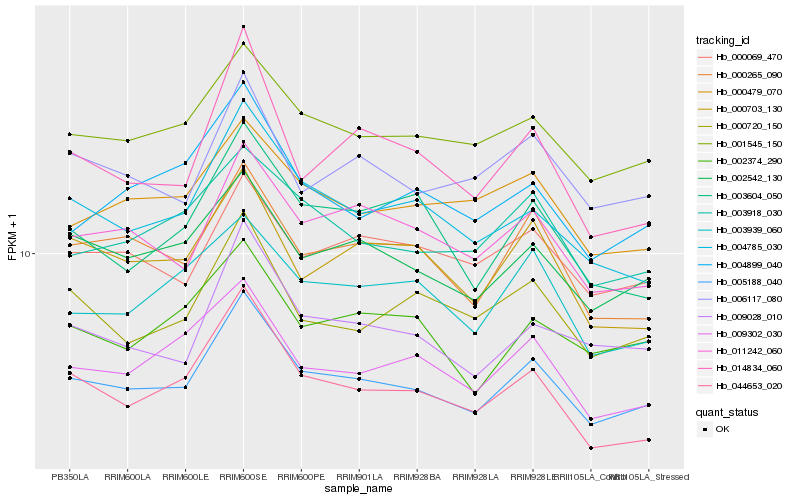
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005188_040 |
0.0 |
- |
- |
PREDICTED: protein furry homolog-like [Nicotiana sylvestris] |
| 2 |
Hb_002542_130 |
0.0658895322 |
transcription factor |
TF Family: HB |
Homeobox protein LUMINIDEPENDENS, putative [Ricinus communis] |
| 3 |
Hb_004785_030 |
0.0676356275 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor VIP1 [Jatropha curcas] |
| 4 |
Hb_001545_150 |
0.0678307352 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105635527 [Jatropha curcas] |
| 5 |
Hb_011242_060 |
0.0695969936 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 6 |
Hb_000265_090 |
0.0696334215 |
- |
- |
PREDICTED: flowering time control protein FPA [Jatropha curcas] |
| 7 |
Hb_009028_010 |
0.0708084073 |
- |
- |
PREDICTED: probable disease resistance protein At1g12280 [Jatropha curcas] |
| 8 |
Hb_044653_020 |
0.0744109875 |
transcription factor, desease resistance |
TF Family: GNAT, Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 9 |
Hb_009302_030 |
0.0783365937 |
- |
- |
hypothetical protein RCOM_1176340 [Ricinus communis] |
| 10 |
Hb_014834_060 |
0.0784331359 |
- |
- |
PREDICTED: uncharacterized protein LOC105632277 isoform X1 [Jatropha curcas] |
| 11 |
Hb_006117_080 |
0.0788163326 |
- |
- |
PREDICTED: transcription factor GTE4 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000703_130 |
0.078897701 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 [Jatropha curcas] |
| 13 |
Hb_000720_150 |
0.0795891253 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X3 [Jatropha curcas] |
| 14 |
Hb_003918_030 |
0.080385022 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 1C-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_000069_470 |
0.0815050833 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 16 |
Hb_003604_050 |
0.0831380987 |
- |
- |
PREDICTED: MAP3K epsilon protein kinase 1-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_002374_290 |
0.0833499118 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 26 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004899_040 |
0.0859253704 |
transcription factor |
TF Family: ARF |
Auxin response factor [Medicago truncatula] |
| 19 |
Hb_003939_060 |
0.0859394046 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000479_070 |
0.0860320657 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |