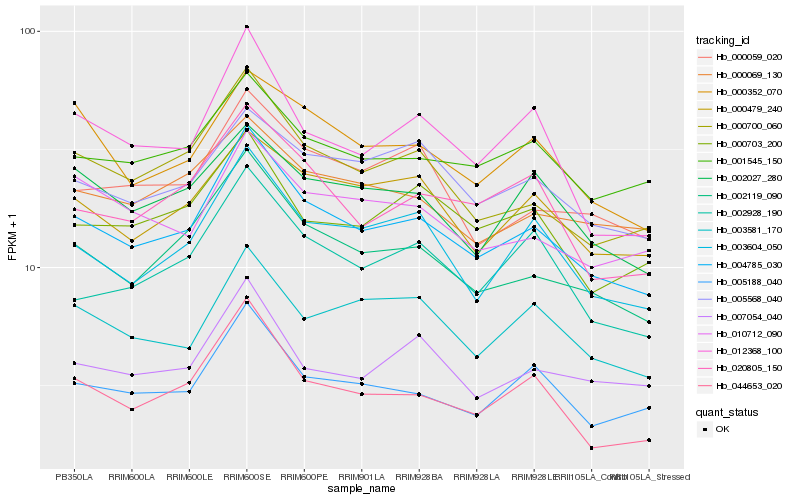
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004785_030 |
0.0 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor VIP1 [Jatropha curcas] |
| 2 |
Hb_002119_090 |
0.0514058851 |
- |
- |
PREDICTED: protein OBERON 3 [Jatropha curcas] |
| 3 |
Hb_000700_060 |
0.0621816656 |
- |
- |
PREDICTED: uncharacterized protein LOC105644416 [Jatropha curcas] |
| 4 |
Hb_005188_040 |
0.0676356275 |
- |
- |
PREDICTED: protein furry homolog-like [Nicotiana sylvestris] |
| 5 |
Hb_044653_020 |
0.0676904042 |
transcription factor, desease resistance |
TF Family: GNAT, Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 6 |
Hb_003604_050 |
0.0677034056 |
- |
- |
PREDICTED: MAP3K epsilon protein kinase 1-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_020805_150 |
0.0750109918 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 8 |
Hb_012368_100 |
0.0761126866 |
- |
- |
PREDICTED: uncharacterized protein LOC105637426 [Jatropha curcas] |
| 9 |
Hb_001545_150 |
0.0763795334 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105635527 [Jatropha curcas] |
| 10 |
Hb_000479_240 |
0.0765773958 |
- |
- |
PREDICTED: probable ubiquitin conjugation factor E4 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000352_070 |
0.0779901257 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 12 |
Hb_000059_020 |
0.0788959667 |
- |
- |
PREDICTED: F-box protein SKP2A [Jatropha curcas] |
| 13 |
Hb_002928_190 |
0.0801910459 |
transcription factor |
TF Family: RWP-RK |
PREDICTED: protein NLP4 [Jatropha curcas] |
| 14 |
Hb_000703_200 |
0.0813122819 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_005568_040 |
0.0813909196 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X2 [Jatropha curcas] |
| 16 |
Hb_007054_040 |
0.0815126431 |
- |
- |
PREDICTED: protein RST1 [Jatropha curcas] |
| 17 |
Hb_002027_280 |
0.0816632547 |
- |
- |
PREDICTED: sister-chromatid cohesion protein 3 [Jatropha curcas] |
| 18 |
Hb_003581_170 |
0.0820324059 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000069_130 |
0.0820754417 |
- |
- |
PREDICTED: uncharacterized protein LOC105642829 isoform X1 [Jatropha curcas] |
| 20 |
Hb_010712_090 |
0.0822861703 |
- |
- |
PREDICTED: uncharacterized protein LOC105631276 [Jatropha curcas] |