| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
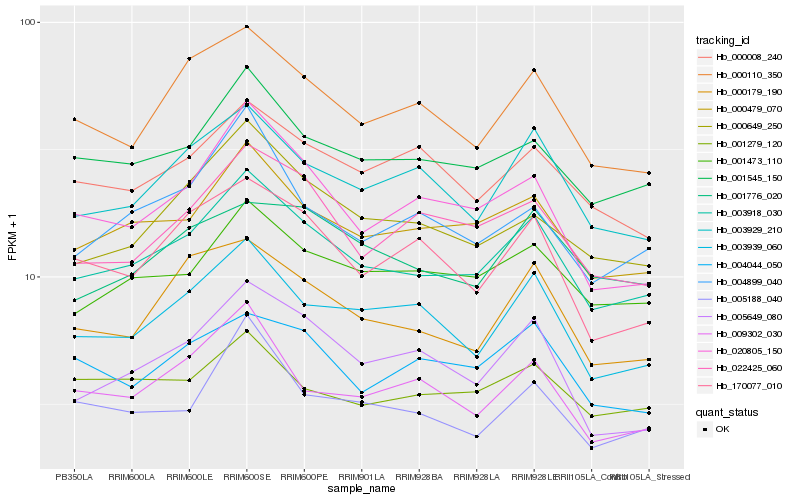
Hb_003918_030 |
0.0 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 1C-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000479_070 |
0.0486958127 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 3 |
Hb_001545_150 |
0.0571042092 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105635527 [Jatropha curcas] |
| 4 |
Hb_001473_110 |
0.0617650238 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 13C-like [Jatropha curcas] |
| 5 |
Hb_003929_210 |
0.0647863096 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 6 |
Hb_003939_060 |
0.065548564 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000110_350 |
0.0656342118 |
- |
- |
PREDICTED: ubiquitin receptor RAD23c-like isoform X2 [Gossypium raimondii] |
| 8 |
Hb_000179_190 |
0.0680086821 |
transcription factor |
TF Family: Alfin-like |
PREDICTED: PHD finger protein ALFIN-LIKE 4 isoform X1 [Vitis vinifera] |
| 9 |
Hb_000649_250 |
0.0703547175 |
transcription factor |
TF Family: C2H2 |
PREDICTED: UBX domain-containing protein 6 [Jatropha curcas] |
| 10 |
Hb_022425_060 |
0.0709032877 |
- |
- |
PREDICTED: protein phosphatase 2C and cyclic nucleotide-binding/kinase domain-containing protein isoform X1 [Jatropha curcas] |
| 11 |
Hb_020805_150 |
0.0720976015 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 12 |
Hb_000008_240 |
0.0774855522 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_170077_010 |
0.0779349357 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001776_020 |
0.0797424623 |
- |
- |
Protein kinase capable of phosphorylating tyrosine family protein [Populus trichocarpa] |
| 15 |
Hb_004899_040 |
0.0799366928 |
transcription factor |
TF Family: ARF |
Auxin response factor [Medicago truncatula] |
| 16 |
Hb_009302_030 |
0.0802293417 |
- |
- |
hypothetical protein RCOM_1176340 [Ricinus communis] |
| 17 |
Hb_005188_040 |
0.080385022 |
- |
- |
PREDICTED: protein furry homolog-like [Nicotiana sylvestris] |
| 18 |
Hb_004044_050 |
0.0804884934 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Jatropha curcas] |
| 19 |
Hb_005649_080 |
0.0807869034 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 20 |
Hb_001279_120 |
0.0817383312 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5-like [Jatropha curcas] |