| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
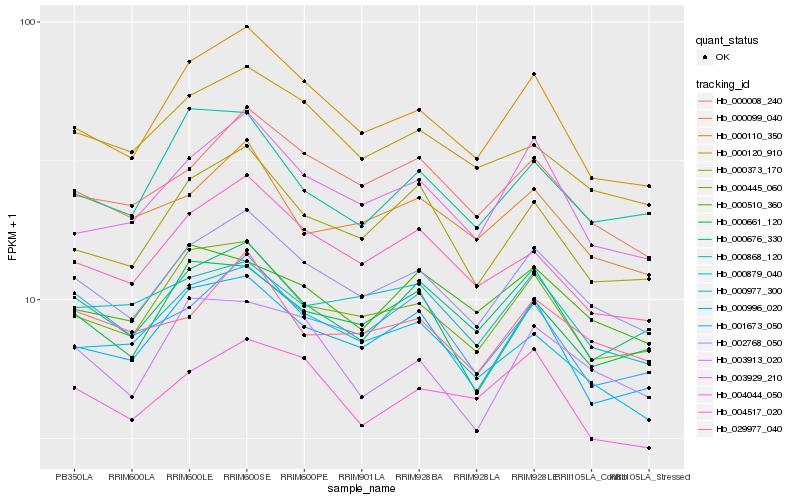
Hb_002768_050 |
0.0 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 2 |
Hb_000110_350 |
0.0506135435 |
- |
- |
PREDICTED: ubiquitin receptor RAD23c-like isoform X2 [Gossypium raimondii] |
| 3 |
Hb_000445_060 |
0.051066567 |
- |
- |
PREDICTED: uncharacterized protein LOC105638996 isoform X3 [Jatropha curcas] |
| 4 |
Hb_000008_240 |
0.0519075428 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000676_330 |
0.0579003328 |
- |
- |
protein with unknown function [Ricinus communis] |
| 6 |
Hb_004517_020 |
0.058972157 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2B isoform X1 [Jatropha curcas] |
| 7 |
Hb_003929_210 |
0.0601327943 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 8 |
Hb_000373_170 |
0.0603477056 |
- |
- |
PREDICTED: exocyst complex component SEC5A-like [Jatropha curcas] |
| 9 |
Hb_000996_020 |
0.0605889096 |
- |
- |
PREDICTED: RNA-binding protein NOB1 [Jatropha curcas] |
| 10 |
Hb_029977_040 |
0.0630461186 |
- |
- |
PREDICTED: uncharacterized protein LOC105640155 isoform X4 [Jatropha curcas] |
| 11 |
Hb_000120_910 |
0.0658357065 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |
| 12 |
Hb_000661_120 |
0.0683604375 |
- |
- |
cap binding protein, putative [Ricinus communis] |
| 13 |
Hb_004044_050 |
0.0706482266 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Jatropha curcas] |
| 14 |
Hb_000099_040 |
0.0712934377 |
- |
- |
unnamed protein product [Coffea canephora] |
| 15 |
Hb_000879_040 |
0.0714577073 |
- |
- |
PREDICTED: calcium-transporting ATPase 3, endoplasmic reticulum-type [Jatropha curcas] |
| 16 |
Hb_001673_050 |
0.0715553872 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_003913_020 |
0.0717347962 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At5g50170 [Jatropha curcas] |
| 18 |
Hb_000977_300 |
0.0720613926 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable E3 ubiquitin-protein ligase ARI8 [Malus domestica] |
| 19 |
Hb_000510_360 |
0.0721093087 |
transcription factor |
TF Family: GNAT |
PREDICTED: histone acetyltransferase GCN5 [Jatropha curcas] |
| 20 |
Hb_000868_120 |
0.0727785018 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 2 [Jatropha curcas] |