| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
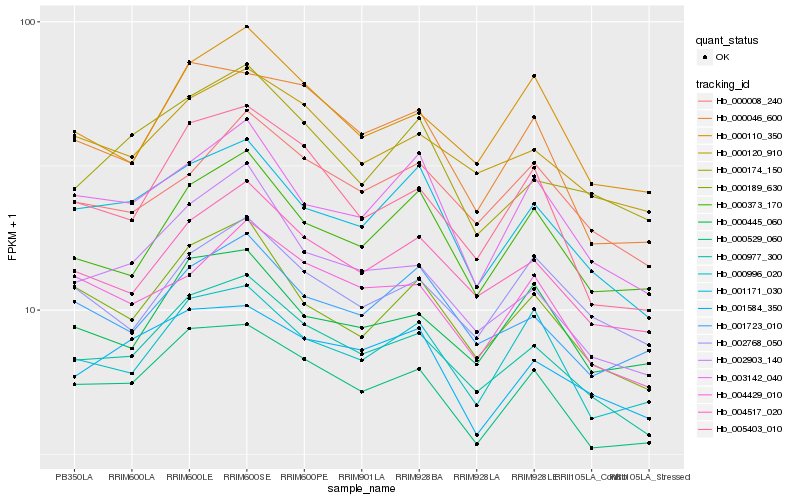
Hb_000977_300 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable E3 ubiquitin-protein ligase ARI8 [Malus domestica] |
| 2 |
Hb_004517_020 |
0.0420468157 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2B isoform X1 [Jatropha curcas] |
| 3 |
Hb_000529_060 |
0.0456294295 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 17 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000120_910 |
0.0484562903 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |
| 5 |
Hb_001171_030 |
0.05788473 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like [Jatropha curcas] |
| 6 |
Hb_000189_630 |
0.0579259656 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 7 |
Hb_000110_350 |
0.0643061947 |
- |
- |
PREDICTED: ubiquitin receptor RAD23c-like isoform X2 [Gossypium raimondii] |
| 8 |
Hb_002903_140 |
0.0648654376 |
- |
- |
PREDICTED: uncharacterized protein LOC105649513 [Jatropha curcas] |
| 9 |
Hb_000008_240 |
0.0659264948 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001584_350 |
0.0666749682 |
- |
- |
hypothetical protein JCGZ_23178 [Jatropha curcas] |
| 11 |
Hb_004429_010 |
0.0669452439 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 12 |
Hb_000046_600 |
0.0675782244 |
- |
- |
PREDICTED: cullin-1-like [Jatropha curcas] |
| 13 |
Hb_005403_010 |
0.0683183795 |
- |
- |
PREDICTED: exocyst complex component EXO70B1-like [Jatropha curcas] |
| 14 |
Hb_000373_170 |
0.0685332814 |
- |
- |
PREDICTED: exocyst complex component SEC5A-like [Jatropha curcas] |
| 15 |
Hb_001723_010 |
0.0688621371 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL6 [Jatropha curcas] |
| 16 |
Hb_000445_060 |
0.0689354719 |
- |
- |
PREDICTED: uncharacterized protein LOC105638996 isoform X3 [Jatropha curcas] |
| 17 |
Hb_000174_150 |
0.0693423048 |
- |
- |
stearoyl-acyl-carrier protein desaturase [Ricinus communis] |
| 18 |
Hb_000996_020 |
0.0694463842 |
- |
- |
PREDICTED: RNA-binding protein NOB1 [Jatropha curcas] |
| 19 |
Hb_003142_040 |
0.0708657124 |
- |
- |
Sf3a3 [Gossypium arboreum] |
| 20 |
Hb_002768_050 |
0.0720613926 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |