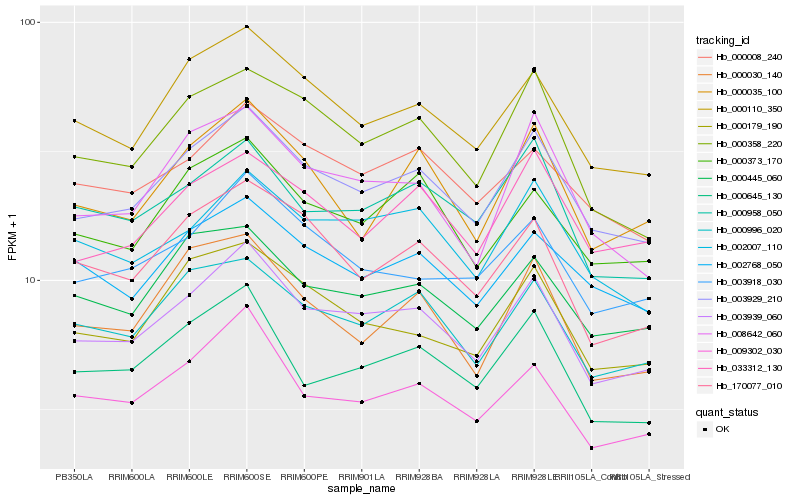
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003929_210 |
0.0 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 2 |
Hb_003939_060 |
0.038948554 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000110_350 |
0.0482613833 |
- |
- |
PREDICTED: ubiquitin receptor RAD23c-like isoform X2 [Gossypium raimondii] |
| 4 |
Hb_033312_130 |
0.0520787202 |
- |
- |
PREDICTED: protein TIC 40, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000996_020 |
0.0571653888 |
- |
- |
PREDICTED: RNA-binding protein NOB1 [Jatropha curcas] |
| 6 |
Hb_000358_220 |
0.0572950312 |
- |
- |
PREDICTED: SNW/SKI-interacting protein-like [Jatropha curcas] |
| 7 |
Hb_002768_050 |
0.0601327943 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 8 |
Hb_000373_170 |
0.0615002699 |
- |
- |
PREDICTED: exocyst complex component SEC5A-like [Jatropha curcas] |
| 9 |
Hb_000008_240 |
0.0616250542 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000445_060 |
0.0618342631 |
- |
- |
PREDICTED: uncharacterized protein LOC105638996 isoform X3 [Jatropha curcas] |
| 11 |
Hb_008642_060 |
0.063271552 |
- |
- |
PREDICTED: dynamin-related protein 1E isoform X2 [Jatropha curcas] |
| 12 |
Hb_000179_190 |
0.0634381053 |
transcription factor |
TF Family: Alfin-like |
PREDICTED: PHD finger protein ALFIN-LIKE 4 isoform X1 [Vitis vinifera] |
| 13 |
Hb_003918_030 |
0.0647863096 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 1C-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_002007_110 |
0.065162978 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000035_100 |
0.0672934563 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 16 |
Hb_170077_010 |
0.0687262507 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000958_050 |
0.0690629783 |
- |
- |
hypothetical protein JCGZ_16781 [Jatropha curcas] |
| 18 |
Hb_000645_130 |
0.0737276705 |
- |
- |
PREDICTED: bromodomain and WD repeat-containing protein 3 [Jatropha curcas] |
| 19 |
Hb_000030_140 |
0.0740993682 |
- |
- |
PREDICTED: general negative regulator of transcription subunit 3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_009302_030 |
0.0746989153 |
- |
- |
hypothetical protein RCOM_1176340 [Ricinus communis] |