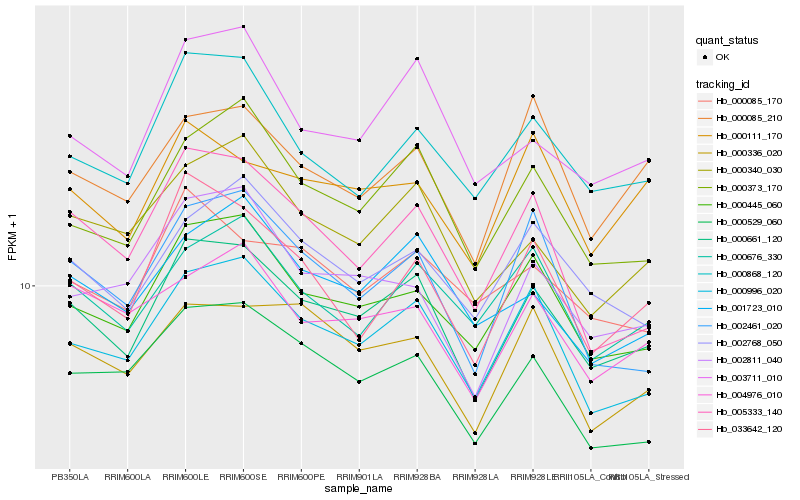
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000661_120 |
0.0 |
- |
- |
cap binding protein, putative [Ricinus communis] |
| 2 |
Hb_000996_020 |
0.0540036946 |
- |
- |
PREDICTED: RNA-binding protein NOB1 [Jatropha curcas] |
| 3 |
Hb_000445_060 |
0.0561468003 |
- |
- |
PREDICTED: uncharacterized protein LOC105638996 isoform X3 [Jatropha curcas] |
| 4 |
Hb_000373_170 |
0.0599684734 |
- |
- |
PREDICTED: exocyst complex component SEC5A-like [Jatropha curcas] |
| 5 |
Hb_000340_030 |
0.0648639418 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle-like [Jatropha curcas] |
| 6 |
Hb_000529_060 |
0.0663608029 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 17 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000868_120 |
0.0665511275 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 2 [Jatropha curcas] |
| 8 |
Hb_003711_010 |
0.0671896238 |
- |
- |
isopropylmalate synthase, putative [Ricinus communis] |
| 9 |
Hb_002768_050 |
0.0683604375 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 10 |
Hb_000336_020 |
0.0690443742 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 2 [Jatropha curcas] |
| 11 |
Hb_001723_010 |
0.0693155058 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL6 [Jatropha curcas] |
| 12 |
Hb_033642_120 |
0.069378892 |
- |
- |
PREDICTED: nucleolar GTP-binding protein 2 [Vitis vinifera] |
| 13 |
Hb_004976_010 |
0.0714473833 |
- |
- |
hypothetical protein RCOM_1429110 [Ricinus communis] |
| 14 |
Hb_000676_330 |
0.072467186 |
- |
- |
protein with unknown function [Ricinus communis] |
| 15 |
Hb_005333_140 |
0.072902294 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 16 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002811_040 |
0.0739206348 |
- |
- |
5'->3' exoribonuclease, putative [Ricinus communis] |
| 17 |
Hb_000085_170 |
0.0745581062 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_002461_020 |
0.0761324574 |
- |
- |
PREDICTED: uncharacterized protein LOC105642649 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000085_210 |
0.0763575793 |
- |
- |
DNA topoisomerase type I, putative [Ricinus communis] |
| 20 |
Hb_000111_170 |
0.0768734729 |
- |
- |
PREDICTED: uncharacterized protein LOC105631234 isoform X1 [Jatropha curcas] |