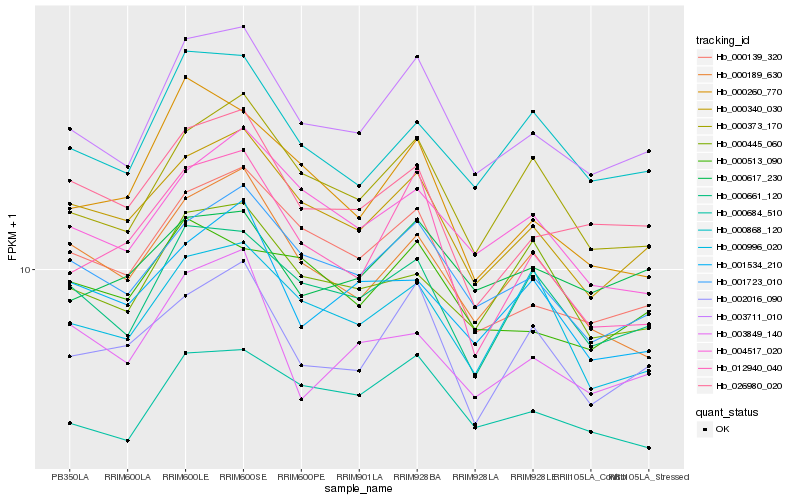
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003711_010 |
0.0 |
- |
- |
isopropylmalate synthase, putative [Ricinus communis] |
| 2 |
Hb_001723_010 |
0.0553384938 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL6 [Jatropha curcas] |
| 3 |
Hb_000340_030 |
0.0597573807 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle-like [Jatropha curcas] |
| 4 |
Hb_000684_510 |
0.0619352441 |
- |
- |
run and tbc1 domain containing 3, plant, putative [Ricinus communis] |
| 5 |
Hb_000373_170 |
0.0619796011 |
- |
- |
PREDICTED: exocyst complex component SEC5A-like [Jatropha curcas] |
| 6 |
Hb_000139_320 |
0.0640108037 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A [Jatropha curcas] |
| 7 |
Hb_000661_120 |
0.0671896238 |
- |
- |
cap binding protein, putative [Ricinus communis] |
| 8 |
Hb_000868_120 |
0.0715177859 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 2 [Jatropha curcas] |
| 9 |
Hb_002016_090 |
0.0729040556 |
- |
- |
PREDICTED: DNA mismatch repair protein PMS1 [Jatropha curcas] |
| 10 |
Hb_026980_020 |
0.0740842497 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0234100-like [Jatropha curcas] |
| 11 |
Hb_004517_020 |
0.0742114884 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2B isoform X1 [Jatropha curcas] |
| 12 |
Hb_000617_230 |
0.0820068281 |
- |
- |
PREDICTED: tafazzin isoform X1 [Jatropha curcas] |
| 13 |
Hb_000189_630 |
0.0827093417 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 14 |
Hb_000513_090 |
0.0832385189 |
- |
- |
PREDICTED: isocitrate dehydrogenase [NADP] [Jatropha curcas] |
| 15 |
Hb_000445_060 |
0.0846866558 |
- |
- |
PREDICTED: uncharacterized protein LOC105638996 isoform X3 [Jatropha curcas] |
| 16 |
Hb_012940_040 |
0.0848956993 |
- |
- |
Splicing factor, arginine/serine-rich, putative [Ricinus communis] |
| 17 |
Hb_003849_140 |
0.085642835 |
- |
- |
PREDICTED: uncharacterized protein LOC105644697 [Jatropha curcas] |
| 18 |
Hb_000996_020 |
0.0869729286 |
- |
- |
PREDICTED: RNA-binding protein NOB1 [Jatropha curcas] |
| 19 |
Hb_000260_770 |
0.0879328062 |
- |
- |
er lumen protein retaining receptor, putative [Ricinus communis] |
| 20 |
Hb_001534_210 |
0.0887806395 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105642316 isoform X2 [Jatropha curcas] |