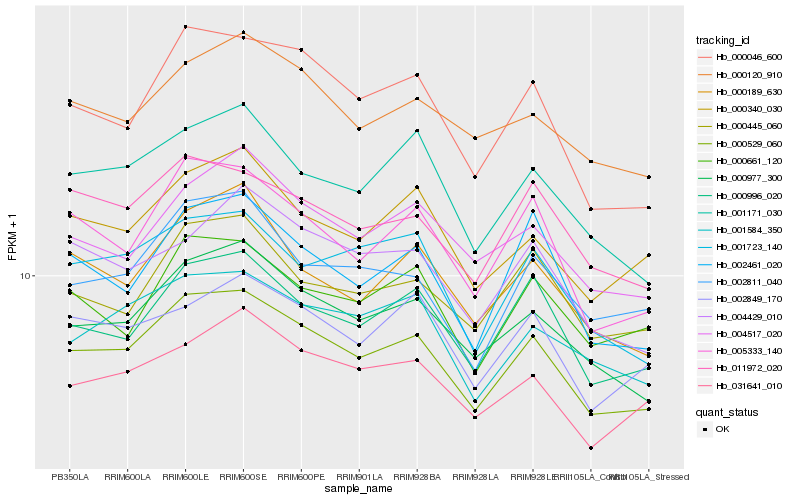
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000529_060 |
0.0 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 17 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000977_300 |
0.0456294295 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable E3 ubiquitin-protein ligase ARI8 [Malus domestica] |
| 3 |
Hb_000996_020 |
0.0525549697 |
- |
- |
PREDICTED: RNA-binding protein NOB1 [Jatropha curcas] |
| 4 |
Hb_000046_600 |
0.0528366469 |
- |
- |
PREDICTED: cullin-1-like [Jatropha curcas] |
| 5 |
Hb_005333_140 |
0.0538103866 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 16 isoform X1 [Jatropha curcas] |
| 6 |
Hb_011972_020 |
0.0573575289 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 7 |
Hb_001171_030 |
0.0589643721 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like [Jatropha curcas] |
| 8 |
Hb_001584_350 |
0.0591144296 |
- |
- |
hypothetical protein JCGZ_23178 [Jatropha curcas] |
| 9 |
Hb_031641_010 |
0.0613354856 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000340_030 |
0.0613748991 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle-like [Jatropha curcas] |
| 11 |
Hb_000120_910 |
0.0616482758 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |
| 12 |
Hb_000445_060 |
0.0619803725 |
- |
- |
PREDICTED: uncharacterized protein LOC105638996 isoform X3 [Jatropha curcas] |
| 13 |
Hb_004517_020 |
0.0620609042 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2B isoform X1 [Jatropha curcas] |
| 14 |
Hb_001723_140 |
0.063320678 |
- |
- |
PREDICTED: importin beta-like SAD2 homolog isoform X2 [Jatropha curcas] |
| 15 |
Hb_000189_630 |
0.064077414 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 16 |
Hb_002811_040 |
0.0644527359 |
- |
- |
5'->3' exoribonuclease, putative [Ricinus communis] |
| 17 |
Hb_002461_020 |
0.0644706747 |
- |
- |
PREDICTED: uncharacterized protein LOC105642649 isoform X2 [Jatropha curcas] |
| 18 |
Hb_002849_170 |
0.0656640509 |
- |
- |
PREDICTED: uncharacterized protein LOC105642955 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004429_010 |
0.0657118105 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 20 |
Hb_000661_120 |
0.0663608029 |
- |
- |
cap binding protein, putative [Ricinus communis] |