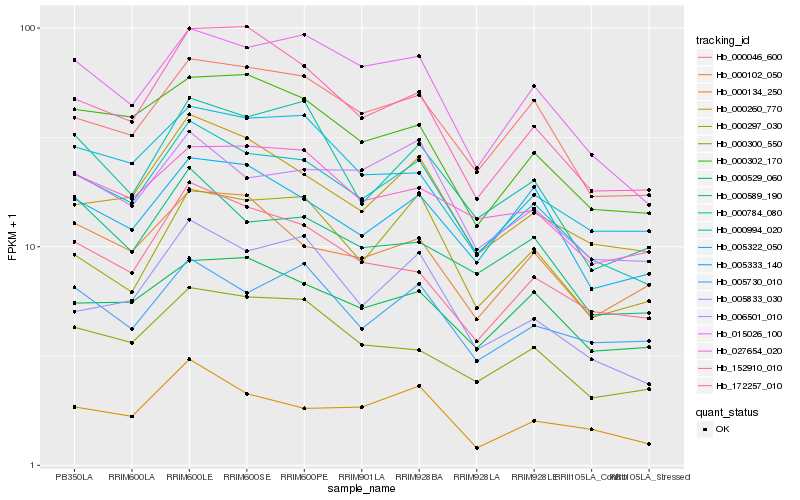
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000994_020 |
0.0 |
- |
- |
methylenetetrahydrofolate dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_015026_100 |
0.0629207447 |
- |
- |
PREDICTED: transmembrane protein 184C-like [Jatropha curcas] |
| 3 |
Hb_000046_600 |
0.0662735827 |
- |
- |
PREDICTED: cullin-1-like [Jatropha curcas] |
| 4 |
Hb_000260_770 |
0.0704313312 |
- |
- |
er lumen protein retaining receptor, putative [Ricinus communis] |
| 5 |
Hb_006501_010 |
0.0751235492 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3A isoform X1 [Populus euphratica] |
| 6 |
Hb_000302_170 |
0.0770470151 |
- |
- |
PREDICTED: dnaJ protein ERDJ2A-like [Jatropha curcas] |
| 7 |
Hb_000300_550 |
0.082750127 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 8 |
Hb_005333_140 |
0.0832896787 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 16 isoform X1 [Jatropha curcas] |
| 9 |
Hb_005322_050 |
0.084161839 |
- |
- |
hypothetical protein POPTR_0009s02400g [Populus trichocarpa] |
| 10 |
Hb_027654_020 |
0.0842186088 |
- |
- |
PREDICTED: LIMR family protein At5g01460 [Jatropha curcas] |
| 11 |
Hb_000529_060 |
0.0846623371 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 17 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000589_190 |
0.0847886742 |
- |
- |
PREDICTED: actin-related protein 5 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000784_080 |
0.0851217635 |
- |
- |
protein disulfide isomerase, putative [Ricinus communis] |
| 14 |
Hb_152910_010 |
0.0856934746 |
- |
- |
PREDICTED: uncharacterized protein LOC105632212 [Jatropha curcas] |
| 15 |
Hb_172257_010 |
0.0857608697 |
transcription factor |
TF Family: HMG |
high mobility group family protein [Populus trichocarpa] |
| 16 |
Hb_000134_250 |
0.0862352115 |
- |
- |
protein with unknown function [Ricinus communis] |
| 17 |
Hb_005833_030 |
0.0865608323 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g16860 [Jatropha curcas] |
| 18 |
Hb_005730_010 |
0.0869633876 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 7 isoform X1 [Populus euphratica] |
| 19 |
Hb_000102_050 |
0.0893736062 |
transcription factor |
TF Family: RB |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000297_030 |
0.0913295381 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |