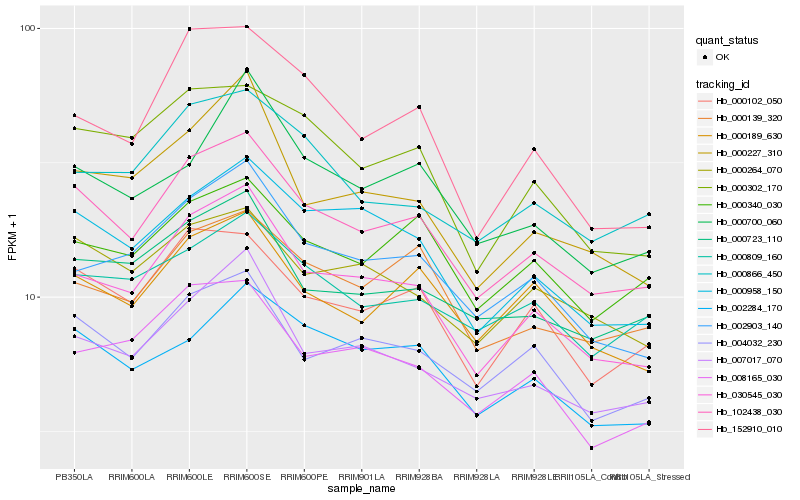
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_102438_030 |
0.0 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 37-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_030545_030 |
0.0525863131 |
- |
- |
PREDICTED: uncharacterized protein LOC105648743 [Jatropha curcas] |
| 3 |
Hb_000102_050 |
0.0578947882 |
transcription factor |
TF Family: RB |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004032_230 |
0.0633157254 |
- |
- |
PREDICTED: uncharacterized protein LOC105635143 [Jatropha curcas] |
| 5 |
Hb_007017_070 |
0.0651848155 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 3 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000958_150 |
0.066889475 |
- |
- |
Actin, putative [Ricinus communis] |
| 7 |
Hb_000139_320 |
0.0699379242 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A [Jatropha curcas] |
| 8 |
Hb_000264_070 |
0.0700153845 |
- |
- |
PREDICTED: uncharacterized protein LOC105649513 [Jatropha curcas] |
| 9 |
Hb_000723_110 |
0.0702706861 |
- |
- |
PREDICTED: TSL-kinase interacting protein 1 [Jatropha curcas] |
| 10 |
Hb_000189_630 |
0.0707927346 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 11 |
Hb_002284_170 |
0.0712664194 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1b [Jatropha curcas] |
| 12 |
Hb_000302_170 |
0.0722989449 |
- |
- |
PREDICTED: dnaJ protein ERDJ2A-like [Jatropha curcas] |
| 13 |
Hb_000866_450 |
0.0746883825 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 14 |
Hb_000340_030 |
0.0752740834 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle-like [Jatropha curcas] |
| 15 |
Hb_002903_140 |
0.0755674057 |
- |
- |
PREDICTED: uncharacterized protein LOC105649513 [Jatropha curcas] |
| 16 |
Hb_000809_160 |
0.0763725413 |
- |
- |
hypothetical protein JCGZ_12355 [Jatropha curcas] |
| 17 |
Hb_008165_030 |
0.0796120044 |
- |
- |
PREDICTED: uncharacterized protein LOC105641288 [Jatropha curcas] |
| 18 |
Hb_152910_010 |
0.0802638623 |
- |
- |
PREDICTED: uncharacterized protein LOC105632212 [Jatropha curcas] |
| 19 |
Hb_000227_310 |
0.0818844323 |
- |
- |
PREDICTED: cyclin-T1-4-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_000700_060 |
0.0824819715 |
- |
- |
PREDICTED: uncharacterized protein LOC105644416 [Jatropha curcas] |