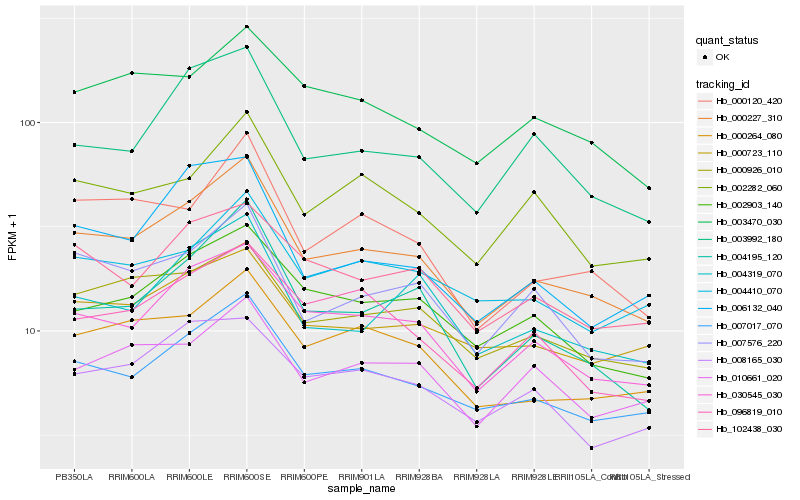
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000227_310 |
0.0 |
- |
- |
PREDICTED: cyclin-T1-4-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_007017_070 |
0.0552599967 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 3 isoform X1 [Jatropha curcas] |
| 3 |
Hb_030545_030 |
0.057753405 |
- |
- |
PREDICTED: uncharacterized protein LOC105648743 [Jatropha curcas] |
| 4 |
Hb_000926_010 |
0.0725318791 |
- |
- |
PREDICTED: uncharacterized protein LOC105632975 [Jatropha curcas] |
| 5 |
Hb_000264_080 |
0.0737918002 |
- |
- |
hypothetical protein JCGZ_21412 [Jatropha curcas] |
| 6 |
Hb_007576_220 |
0.0748133246 |
- |
- |
PREDICTED: kanadaptin [Jatropha curcas] |
| 7 |
Hb_004195_120 |
0.0750695872 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 8 |
Hb_000120_420 |
0.0751629509 |
- |
- |
PREDICTED: AT-rich interactive domain-containing protein 5-like [Jatropha curcas] |
| 9 |
Hb_002903_140 |
0.0758392315 |
- |
- |
PREDICTED: uncharacterized protein LOC105649513 [Jatropha curcas] |
| 10 |
Hb_102438_030 |
0.0818844323 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 37-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_004319_070 |
0.0829396001 |
- |
- |
hypothetical protein RCOM_0852460 [Ricinus communis] |
| 12 |
Hb_003992_180 |
0.083202399 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000723_110 |
0.0837391647 |
- |
- |
PREDICTED: TSL-kinase interacting protein 1 [Jatropha curcas] |
| 14 |
Hb_003470_030 |
0.0846645818 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF126-like [Jatropha curcas] |
| 15 |
Hb_002282_060 |
0.0854852752 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_096819_010 |
0.0860680192 |
- |
- |
PREDICTED: GRIP and coiled-coil domain-containing protein 2 isoform X4 [Jatropha curcas] |
| 17 |
Hb_004410_070 |
0.087716032 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |
| 18 |
Hb_006132_040 |
0.0881188258 |
- |
- |
hypothetical protein JCGZ_14390 [Jatropha curcas] |
| 19 |
Hb_010661_020 |
0.088893366 |
- |
- |
PREDICTED: uncharacterized protein LOC105630767 isoform X1 [Jatropha curcas] |
| 20 |
Hb_008165_030 |
0.089433314 |
- |
- |
PREDICTED: uncharacterized protein LOC105641288 [Jatropha curcas] |