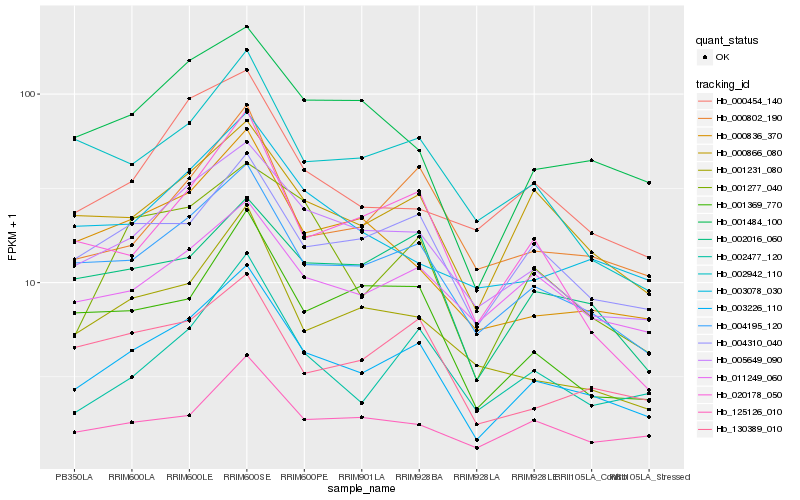
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003226_110 |
0.0 |
- |
- |
PREDICTED: exocyst complex component SEC15B [Jatropha curcas] |
| 2 |
Hb_004195_120 |
0.0846744445 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 3 |
Hb_005649_090 |
0.094822668 |
- |
- |
PREDICTED: ninja-family protein AFP3-like [Jatropha curcas] |
| 4 |
Hb_004310_040 |
0.1106524454 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000866_080 |
0.1140961626 |
- |
- |
PREDICTED: signal peptide peptidase-like 4 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001484_100 |
0.1211924165 |
- |
- |
PREDICTED: cyanate hydratase [Jatropha curcas] |
| 7 |
Hb_002477_120 |
0.1221111659 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 8 |
Hb_000802_190 |
0.1233454437 |
- |
- |
PREDICTED: peroxisomal (S)-2-hydroxy-acid oxidase [Jatropha curcas] |
| 9 |
Hb_001277_040 |
0.1249508037 |
- |
- |
PREDICTED: uncharacterized protein LOC105632066 [Jatropha curcas] |
| 10 |
Hb_130389_010 |
0.1281296276 |
- |
- |
PREDICTED: la protein 1 [Jatropha curcas] |
| 11 |
Hb_000454_140 |
0.1313266368 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2a-like [Jatropha curcas] |
| 12 |
Hb_002016_060 |
0.131714186 |
- |
- |
PREDICTED: uncharacterized protein LOC105642502 [Jatropha curcas] |
| 13 |
Hb_000836_370 |
0.1318405006 |
- |
- |
PREDICTED: protein REVEILLE 3-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_001369_770 |
0.1318673322 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 6-like [Jatropha curcas] |
| 15 |
Hb_001231_080 |
0.1319064253 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER-like isoform X1 [Fragaria vesca subsp. vesca] |
| 16 |
Hb_125126_010 |
0.1324343706 |
desease resistance |
Gene Name: NB-ARC |
resistance family protein [Populus trichocarpa] |
| 17 |
Hb_002942_110 |
0.1337261538 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 2-like [Jatropha curcas] |
| 18 |
Hb_020178_050 |
0.133731074 |
desease resistance |
Gene Name: RPW8 |
PREDICTED: probable disease resistance protein At4g33300 [Jatropha curcas] |
| 19 |
Hb_011249_060 |
0.1343850998 |
- |
- |
hypothetical protein POPTR_0018s12600g [Populus trichocarpa] |
| 20 |
Hb_003078_030 |
0.1350329136 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CHIP [Jatropha curcas] |