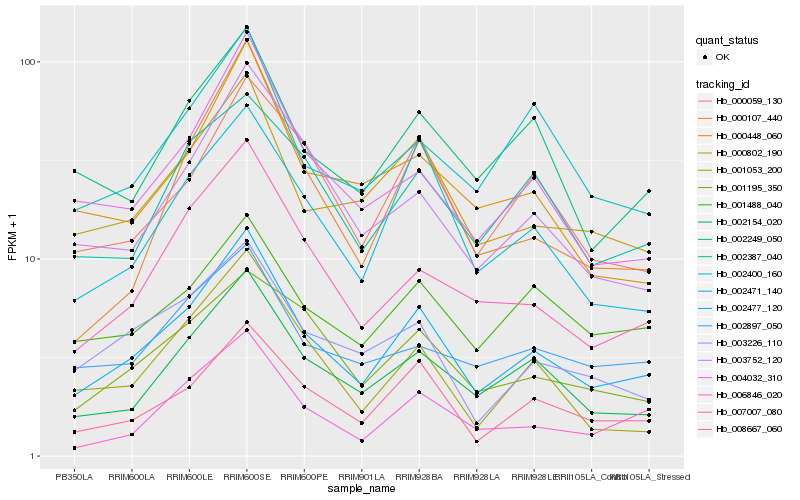
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002477_120 |
0.0 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 2 |
Hb_008667_060 |
0.0912411203 |
- |
- |
Magnesium and cobalt efflux protein corC, putative [Ricinus communis] |
| 3 |
Hb_002154_020 |
0.0950629479 |
- |
- |
protein with unknown function [Ricinus communis] |
| 4 |
Hb_000802_190 |
0.1054981339 |
- |
- |
PREDICTED: peroxisomal (S)-2-hydroxy-acid oxidase [Jatropha curcas] |
| 5 |
Hb_007007_080 |
0.1096980532 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004032_310 |
0.1104554032 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 14 [Jatropha curcas] |
| 7 |
Hb_002471_140 |
0.114247797 |
- |
- |
PREDICTED: protein OBERON 3 [Jatropha curcas] |
| 8 |
Hb_000059_130 |
0.1162164102 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 1 [Jatropha curcas] |
| 9 |
Hb_002249_050 |
0.1188049911 |
- |
- |
Auxin response factor, putative [Ricinus communis] |
| 10 |
Hb_006846_020 |
0.1204342913 |
- |
- |
PREDICTED: uncharacterized protein LOC105648685 [Jatropha curcas] |
| 11 |
Hb_003226_110 |
0.1221111659 |
- |
- |
PREDICTED: exocyst complex component SEC15B [Jatropha curcas] |
| 12 |
Hb_001195_350 |
0.1228410858 |
- |
- |
hypothetical protein JCGZ_04423 [Jatropha curcas] |
| 13 |
Hb_003752_120 |
0.1234775252 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002387_040 |
0.1250457102 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 15 |
Hb_001488_040 |
0.1256250951 |
- |
- |
PREDICTED: nipped-B-like protein A [Jatropha curcas] |
| 16 |
Hb_000448_060 |
0.1275789694 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 17 |
Hb_002897_050 |
0.1281553726 |
- |
- |
PREDICTED: uncharacterized protein LOC105649158 [Jatropha curcas] |
| 18 |
Hb_002400_160 |
0.1297451268 |
- |
- |
Ubiquitin-protein ligase BRE1A, putative [Ricinus communis] |
| 19 |
Hb_001053_200 |
0.132087052 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 20 |
Hb_000107_440 |
0.1330404443 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A synthase |
hydroxymethylglutaryl-CoA reductase [Hevea brasiliensis] |