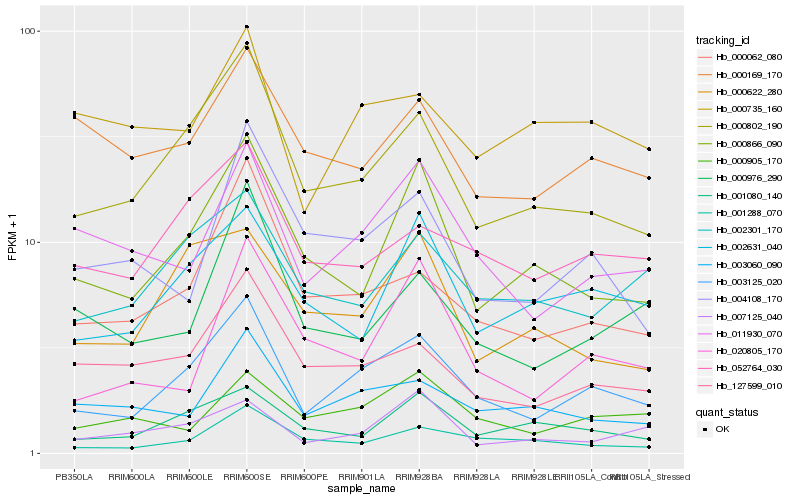
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003125_020 |
0.0 |
- |
- |
AMP-activated protein kinase, gamma regulatory subunit, putative [Ricinus communis] |
| 2 |
Hb_000802_190 |
0.1189669416 |
- |
- |
PREDICTED: peroxisomal (S)-2-hydroxy-acid oxidase [Jatropha curcas] |
| 3 |
Hb_000062_080 |
0.1497159677 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR11-like [Jatropha curcas] |
| 4 |
Hb_011930_070 |
0.1559983202 |
- |
- |
PREDICTED: probable AMP deaminase [Jatropha curcas] |
| 5 |
Hb_052764_030 |
0.1581552351 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os07g0563300 [Jatropha curcas] |
| 6 |
Hb_000866_090 |
0.1620724648 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like isoform X3 [Jatropha curcas] |
| 7 |
Hb_127599_010 |
0.1625722187 |
- |
- |
hypothetical protein CISIN_1g0010462mg, partial [Citrus sinensis] |
| 8 |
Hb_001288_070 |
0.1639442809 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 9 |
Hb_002631_040 |
0.1670207543 |
- |
- |
hypothetical protein EUGRSUZ_J03178 [Eucalyptus grandis] |
| 10 |
Hb_003060_090 |
0.1679471718 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 11 |
Hb_002301_170 |
0.168909828 |
- |
- |
MAC/Perforin domain-containing protein [Theobroma cacao] |
| 12 |
Hb_001080_140 |
0.1738964955 |
transcription factor |
TF Family: RWP-RK |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000905_170 |
0.1755702752 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 14 |
Hb_000976_290 |
0.1778094691 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105646228 [Jatropha curcas] |
| 15 |
Hb_020805_170 |
0.1791365797 |
- |
- |
PREDICTED: F-box protein CPR30-like [Jatropha curcas] |
| 16 |
Hb_004108_170 |
0.1797212671 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000622_280 |
0.1800126125 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000169_170 |
0.1865904242 |
- |
- |
3-hydroxyacyl-CoA dehyrogenase, putative [Ricinus communis] |
| 19 |
Hb_007125_040 |
0.1879679119 |
- |
- |
PREDICTED: cationic amino acid transporter 1-like [Jatropha curcas] |
| 20 |
Hb_000735_160 |
0.1885156305 |
- |
- |
PREDICTED: uncharacterized protein LOC105644513 [Jatropha curcas] |