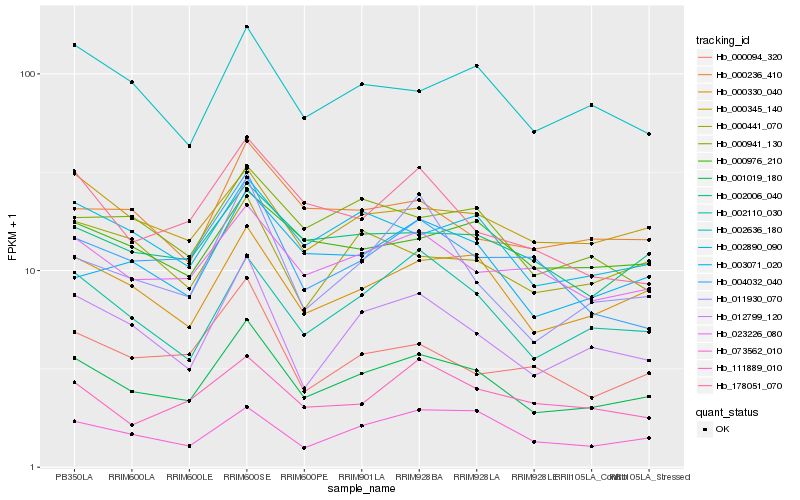
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001019_180 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639324 [Jatropha curcas] |
| 2 |
Hb_000330_040 |
0.0717177549 |
- |
- |
PREDICTED: protein RER1B-like [Jatropha curcas] |
| 3 |
Hb_012799_120 |
0.0852919886 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH3 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002110_030 |
0.0887942027 |
- |
- |
PREDICTED: WD and tetratricopeptide repeats protein 1 isoform X3 [Jatropha curcas] |
| 5 |
Hb_002890_090 |
0.0895604017 |
- |
- |
Auxin response factor 1 [Glycine soja] |
| 6 |
Hb_023226_080 |
0.0907594626 |
transcription factor |
TF Family: C3H |
PREDICTED: 30-kDa cleavage and polyadenylation specificity factor 30 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002006_040 |
0.096891966 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit delta isoform X1 [Jatropha curcas] |
| 8 |
Hb_003071_020 |
0.0988998134 |
transcription factor |
TF Family: ARF |
Auxin response factor [Medicago truncatula] |
| 9 |
Hb_073562_010 |
0.0997852969 |
- |
- |
hypothetical protein POPTR_0011s036602g, partial [Populus trichocarpa] |
| 10 |
Hb_000345_140 |
0.1016117735 |
- |
- |
PREDICTED: autophagy-related protein 18a-like [Jatropha curcas] |
| 11 |
Hb_002636_180 |
0.1024646327 |
- |
- |
inositol or phosphatidylinositol kinase, putative [Ricinus communis] |
| 12 |
Hb_000976_210 |
0.1062387816 |
- |
- |
PREDICTED: protein PAT1 homolog 1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000236_410 |
0.1073527553 |
- |
- |
Paramyosin, putative [Ricinus communis] |
| 14 |
Hb_000941_130 |
0.1084240565 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 15 |
Hb_178051_070 |
0.1093657743 |
- |
- |
PREDICTED: exocyst complex component EXO70B1-like [Jatropha curcas] |
| 16 |
Hb_000094_320 |
0.1106237783 |
- |
- |
hypothetical protein JCGZ_04407 [Jatropha curcas] |
| 17 |
Hb_011930_070 |
0.1107158327 |
- |
- |
PREDICTED: probable AMP deaminase [Jatropha curcas] |
| 18 |
Hb_004032_040 |
0.1117742715 |
- |
- |
unknown [Medicago truncatula] |
| 19 |
Hb_111889_010 |
0.1122451135 |
- |
- |
PREDICTED: DNA ligase 4 [Jatropha curcas] |
| 20 |
Hb_000441_070 |
0.1122790053 |
transcription factor |
TF Family: C3H |
PREDICTED: 30-kDa cleavage and polyadenylation specificity factor 30 isoform X1 [Jatropha curcas] |