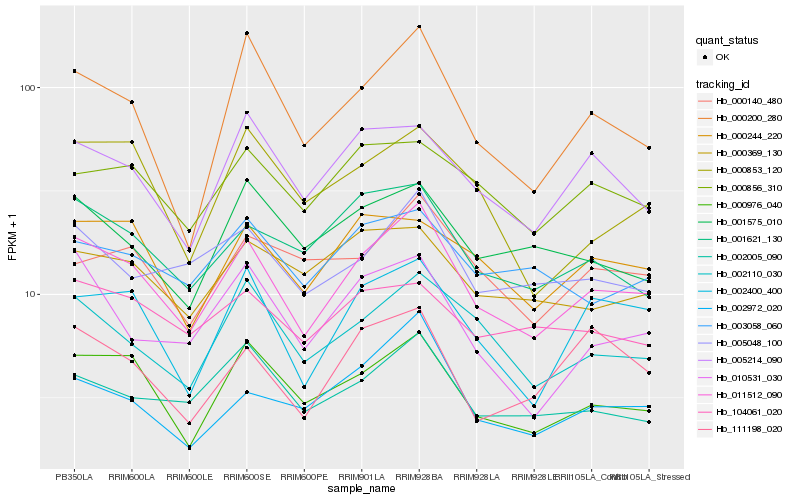
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011512_090 |
0.0 |
- |
- |
Hydroxyethylthiazole kinase, putative [Ricinus communis] |
| 2 |
Hb_000976_040 |
0.0863360033 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 3 |
Hb_002110_030 |
0.0955808593 |
- |
- |
PREDICTED: WD and tetratricopeptide repeats protein 1 isoform X3 [Jatropha curcas] |
| 4 |
Hb_002400_400 |
0.0996834184 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X3 [Jatropha curcas] |
| 5 |
Hb_000200_280 |
0.1014353546 |
- |
- |
PREDICTED: uncharacterized protein LOC105636924 [Jatropha curcas] |
| 6 |
Hb_002005_090 |
0.1032614874 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001621_130 |
0.1054830755 |
- |
- |
PREDICTED: pumilio homolog 1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_005048_100 |
0.106706127 |
- |
- |
secretory carrier membrane protein, putative [Ricinus communis] |
| 9 |
Hb_005214_090 |
0.1087249232 |
- |
- |
PREDICTED: uncharacterized protein LOC105636015 [Jatropha curcas] |
| 10 |
Hb_000244_220 |
0.109521917 |
- |
- |
PREDICTED: bifunctional aspartokinase/homoserine dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 11 |
Hb_002972_020 |
0.1113328648 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 12 |
Hb_000856_310 |
0.1133079005 |
- |
- |
PREDICTED: hypersensitive-induced response protein 1 [Jatropha curcas] |
| 13 |
Hb_010531_030 |
0.1165957999 |
- |
- |
PREDICTED: uncharacterized protein LOC105629654 [Jatropha curcas] |
| 14 |
Hb_000853_120 |
0.1166104682 |
- |
- |
PREDICTED: ethylene response sensor 1 [Jatropha curcas] |
| 15 |
Hb_001575_010 |
0.1176275864 |
- |
- |
PREDICTED: pumilio homolog 6, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_000140_480 |
0.1184362453 |
- |
- |
PREDICTED: uncharacterized protein LOC105637350 [Jatropha curcas] |
| 17 |
Hb_003058_060 |
0.1189067239 |
- |
- |
PREDICTED: proteasome-associated protein ECM29 homolog [Jatropha curcas] |
| 18 |
Hb_111198_020 |
0.1195235373 |
- |
- |
hypothetical protein JCGZ_06907 [Jatropha curcas] |
| 19 |
Hb_000369_130 |
0.119992755 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 4 [Jatropha curcas] |
| 20 |
Hb_104061_020 |
0.1210648475 |
- |
- |
PREDICTED: UV-stimulated scaffold protein A homolog [Jatropha curcas] |