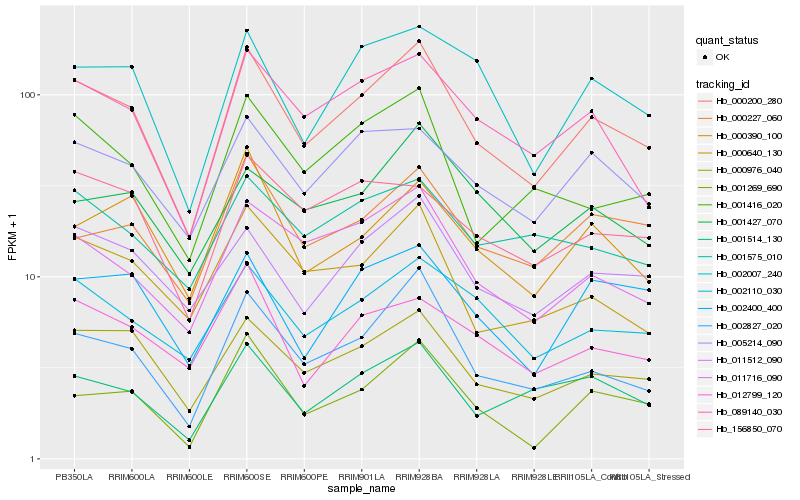
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000200_280 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636924 [Jatropha curcas] |
| 2 |
Hb_000976_040 |
0.0829968564 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 3 |
Hb_002827_020 |
0.0863068847 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 5-like [Jatropha curcas] |
| 4 |
Hb_089140_030 |
0.0966815785 |
- |
- |
PREDICTED: thiol protease aleurain-like [Jatropha curcas] |
| 5 |
Hb_011716_090 |
0.1005865143 |
transcription factor |
TF Family: HRT |
putative transciption factor family protein [Populus trichocarpa] |
| 6 |
Hb_011512_090 |
0.1014353546 |
- |
- |
Hydroxyethylthiazole kinase, putative [Ricinus communis] |
| 7 |
Hb_002110_030 |
0.1051934352 |
- |
- |
PREDICTED: WD and tetratricopeptide repeats protein 1 isoform X3 [Jatropha curcas] |
| 8 |
Hb_005214_090 |
0.105838277 |
- |
- |
PREDICTED: uncharacterized protein LOC105636015 [Jatropha curcas] |
| 9 |
Hb_001514_130 |
0.1077155953 |
- |
- |
hypothetical protein JCGZ_16073 [Jatropha curcas] |
| 10 |
Hb_000640_130 |
0.1083670258 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_002007_240 |
0.1118004721 |
- |
- |
PREDICTED: F-box protein At1g55000 [Jatropha curcas] |
| 12 |
Hb_001416_020 |
0.1130030851 |
- |
- |
H(\+)-transporting atpase plant/fungi plasma membrane type, putative [Ricinus communis] |
| 13 |
Hb_000227_060 |
0.1132840703 |
transcription factor |
TF Family: NF-YC |
hypothetical protein POPTR_0010s03730g [Populus trichocarpa] |
| 14 |
Hb_001269_690 |
0.1139264269 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002400_400 |
0.1183949828 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X3 [Jatropha curcas] |
| 16 |
Hb_156850_070 |
0.1214990621 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000390_100 |
0.1215180117 |
- |
- |
alpha-hydroxynitrile lyase family protein [Populus trichocarpa] |
| 18 |
Hb_001575_010 |
0.1225216137 |
- |
- |
PREDICTED: pumilio homolog 6, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_012799_120 |
0.1236476879 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001427_070 |
0.1241518126 |
- |
- |
PREDICTED: xaa-Pro dipeptidase [Jatropha curcas] |