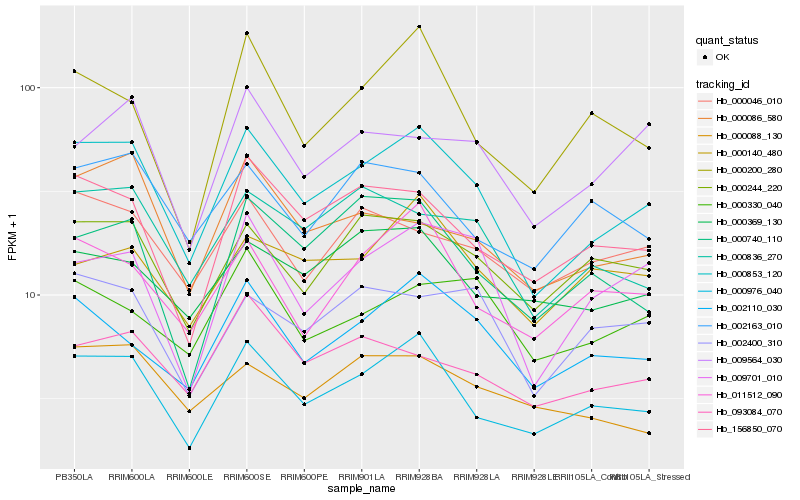
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000853_120 |
0.0 |
- |
- |
PREDICTED: ethylene response sensor 1 [Jatropha curcas] |
| 2 |
Hb_000976_040 |
0.0801972594 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 3 |
Hb_002110_030 |
0.1079179169 |
- |
- |
PREDICTED: WD and tetratricopeptide repeats protein 1 isoform X3 [Jatropha curcas] |
| 4 |
Hb_156850_070 |
0.1090176004 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000836_270 |
0.1103750453 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog MMK2 [Jatropha curcas] |
| 6 |
Hb_000244_220 |
0.1119515509 |
- |
- |
PREDICTED: bifunctional aspartokinase/homoserine dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_009701_010 |
0.1139636901 |
- |
- |
PREDICTED: putative inactive cadmium/zinc-transporting ATPase HMA3 [Jatropha curcas] |
| 8 |
Hb_011512_090 |
0.1166104682 |
- |
- |
Hydroxyethylthiazole kinase, putative [Ricinus communis] |
| 9 |
Hb_002400_310 |
0.1189724427 |
- |
- |
hypothetical protein JCGZ_07138 [Jatropha curcas] |
| 10 |
Hb_000046_010 |
0.1219210094 |
- |
- |
PREDICTED: histidine-containing phosphotransfer protein 1 [Jatropha curcas] |
| 11 |
Hb_000086_580 |
0.123657545 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 12 |
Hb_093084_070 |
0.1263859923 |
desease resistance |
Gene Name: Baculo_PEP_C |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 13 |
Hb_000200_280 |
0.1274928659 |
- |
- |
PREDICTED: uncharacterized protein LOC105636924 [Jatropha curcas] |
| 14 |
Hb_000740_110 |
0.1278797486 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01850 [Jatropha curcas] |
| 15 |
Hb_002163_010 |
0.1279056561 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410 [Jatropha curcas] |
| 16 |
Hb_000140_480 |
0.1284486112 |
- |
- |
PREDICTED: uncharacterized protein LOC105637350 [Jatropha curcas] |
| 17 |
Hb_000330_040 |
0.1287121358 |
- |
- |
PREDICTED: protein RER1B-like [Jatropha curcas] |
| 18 |
Hb_009564_030 |
0.128911801 |
transcription factor |
TF Family: bHLH |
Transcription factor AtMYC2, putative [Ricinus communis] |
| 19 |
Hb_000369_130 |
0.1289293775 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 4 [Jatropha curcas] |
| 20 |
Hb_000088_130 |
0.1292564036 |
- |
- |
hypothetical protein JCGZ_02447 [Jatropha curcas] |