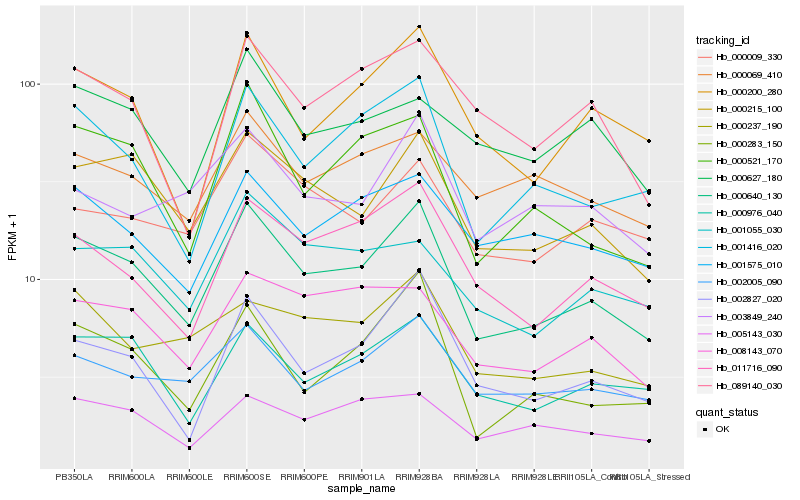
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000640_130 |
0.0 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_000215_100 |
0.0848822906 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001416_020 |
0.0992537801 |
- |
- |
H(\+)-transporting atpase plant/fungi plasma membrane type, putative [Ricinus communis] |
| 4 |
Hb_000976_040 |
0.0998064644 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 5 |
Hb_002005_090 |
0.1047559592 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_011716_090 |
0.1058072761 |
transcription factor |
TF Family: HRT |
putative transciption factor family protein [Populus trichocarpa] |
| 7 |
Hb_000521_170 |
0.1059568728 |
- |
- |
PREDICTED: callose synthase 1 [Jatropha curcas] |
| 8 |
Hb_000200_280 |
0.1083670258 |
- |
- |
PREDICTED: uncharacterized protein LOC105636924 [Jatropha curcas] |
| 9 |
Hb_002827_020 |
0.1125168975 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 5-like [Jatropha curcas] |
| 10 |
Hb_000283_150 |
0.1168948291 |
- |
- |
Ankyrin repeat family protein [Theobroma cacao] |
| 11 |
Hb_008143_070 |
0.1188647085 |
- |
- |
PREDICTED: factor of DNA methylation 4 [Jatropha curcas] |
| 12 |
Hb_000237_190 |
0.1189448644 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 3 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001055_030 |
0.121821197 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 14 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001575_010 |
0.1221578912 |
- |
- |
PREDICTED: pumilio homolog 6, chloroplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_000009_330 |
0.1240034012 |
transcription factor |
TF Family: LIM |
zinc ion binding protein, putative [Ricinus communis] |
| 16 |
Hb_005143_030 |
0.124024042 |
- |
- |
PREDICTED: uncharacterized protein LOC105638843 [Jatropha curcas] |
| 17 |
Hb_089140_030 |
0.1258286842 |
- |
- |
PREDICTED: thiol protease aleurain-like [Jatropha curcas] |
| 18 |
Hb_000069_410 |
0.1277310276 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 19 |
Hb_000627_180 |
0.1301676041 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003849_240 |
0.1303854391 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |