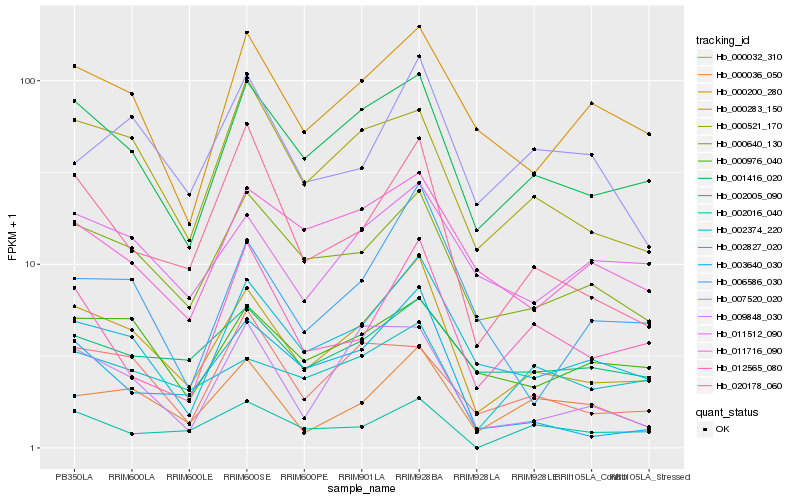
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000283_150 |
0.0 |
- |
- |
Ankyrin repeat family protein [Theobroma cacao] |
| 2 |
Hb_001416_020 |
0.1072150874 |
- |
- |
H(\+)-transporting atpase plant/fungi plasma membrane type, putative [Ricinus communis] |
| 3 |
Hb_000640_130 |
0.1168948291 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_002827_020 |
0.1207025968 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 5-like [Jatropha curcas] |
| 5 |
Hb_000521_170 |
0.1379344886 |
- |
- |
PREDICTED: callose synthase 1 [Jatropha curcas] |
| 6 |
Hb_000036_050 |
0.1410156354 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 6-like [Camelina sativa] |
| 7 |
Hb_020178_060 |
0.1414223145 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 3 [Jatropha curcas] |
| 8 |
Hb_000200_280 |
0.1520678624 |
- |
- |
PREDICTED: uncharacterized protein LOC105636924 [Jatropha curcas] |
| 9 |
Hb_000976_040 |
0.1526809151 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 10 |
Hb_011512_090 |
0.1590581337 |
- |
- |
Hydroxyethylthiazole kinase, putative [Ricinus communis] |
| 11 |
Hb_002005_090 |
0.1607246399 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_006586_030 |
0.1649207486 |
- |
- |
PREDICTED: transcription factor LHW isoform X1 [Jatropha curcas] |
| 13 |
Hb_003640_030 |
0.1649354996 |
- |
- |
PREDICTED: uncharacterized protein LOC105647345 isoform X1 [Jatropha curcas] |
| 14 |
Hb_011716_090 |
0.1683321159 |
transcription factor |
TF Family: HRT |
putative transciption factor family protein [Populus trichocarpa] |
| 15 |
Hb_002374_220 |
0.1685438771 |
- |
- |
hypothetical protein CICLE_v10001597mg [Citrus clementina] |
| 16 |
Hb_000032_310 |
0.1701942392 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 17 |
Hb_012565_080 |
0.1702129762 |
- |
- |
serine/threonine kinase, putative [Ricinus communis] |
| 18 |
Hb_007520_020 |
0.1709866252 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose 5-phosphate reductoisomerase |
1-deoxy-D-xylulose-5-phosphate reductoisomerase [Hevea brasiliensis] |
| 19 |
Hb_009848_030 |
0.1710473011 |
- |
- |
long-chain-fatty-acid CoA ligase, putative [Ricinus communis] |
| 20 |
Hb_002016_040 |
0.1732505331 |
rubber biosynthesis |
Gene Name: SRPP8 |
PREDICTED: stress-related protein [Jatropha curcas] |