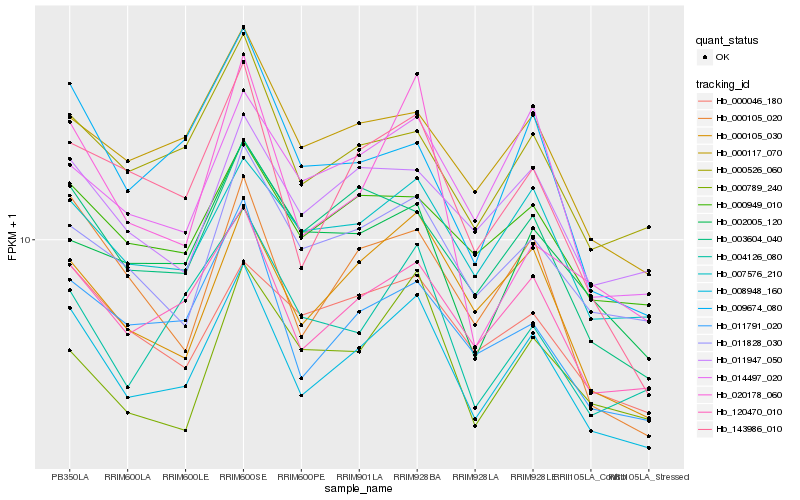
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008948_160 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104214688 isoform X2 [Nicotiana sylvestris] |
| 2 |
Hb_120470_010 |
0.0994651852 |
- |
- |
PREDICTED: PAX3- and PAX7-binding protein 1 [Jatropha curcas] |
| 3 |
Hb_000105_030 |
0.1088468435 |
- |
- |
fkbp-rapamycin associated protein, putative [Ricinus communis] |
| 4 |
Hb_143986_010 |
0.1090549698 |
- |
- |
PREDICTED: beta-galactosidase [Jatropha curcas] |
| 5 |
Hb_000105_020 |
0.1150622945 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOR-like [Malus domestica] |
| 6 |
Hb_011791_020 |
0.1252789281 |
- |
- |
hypothetical protein Csa_6G486930 [Cucumis sativus] |
| 7 |
Hb_003604_040 |
0.129693003 |
- |
- |
Serine/threonine-protein kinase sepA [Morus notabilis] |
| 8 |
Hb_009674_080 |
0.1357869211 |
- |
- |
PREDICTED: branchpoint-bridging protein [Jatropha curcas] |
| 9 |
Hb_007576_210 |
0.1374712716 |
- |
- |
PREDICTED: protein furry homolog [Jatropha curcas] |
| 10 |
Hb_000117_070 |
0.1392847708 |
- |
- |
PREDICTED: branchpoint-bridging protein [Jatropha curcas] |
| 11 |
Hb_011828_030 |
0.141538687 |
- |
- |
ubiquitin protein ligase E3a, putative [Ricinus communis] |
| 12 |
Hb_000526_060 |
0.141684397 |
- |
- |
PREDICTED: clustered mitochondria protein [Jatropha curcas] |
| 13 |
Hb_014497_020 |
0.1419635196 |
- |
- |
PREDICTED: U5 small nuclear ribonucleoprotein 200 kDa helicase [Jatropha curcas] |
| 14 |
Hb_020178_060 |
0.1424569005 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 3 [Jatropha curcas] |
| 15 |
Hb_004126_080 |
0.1447938764 |
- |
- |
mitochondrial 50S ribosomal protein L21 [Hevea brasiliensis] |
| 16 |
Hb_000046_180 |
0.1489451507 |
- |
- |
PREDICTED: uncharacterized protein LOC105631840 [Jatropha curcas] |
| 17 |
Hb_011947_050 |
0.1513835938 |
- |
- |
E3 ubiquitin protein ligase upl2, putative [Ricinus communis] |
| 18 |
Hb_000949_010 |
0.1515051843 |
- |
- |
PREDICTED: regulator of nonsense transcripts UPF2 [Jatropha curcas] |
| 19 |
Hb_002005_120 |
0.1516783668 |
- |
- |
PREDICTED: protein MEI2-like 5 [Jatropha curcas] |
| 20 |
Hb_000789_240 |
0.1525356377 |
- |
- |
gamma glutamyl transpeptidases, putative [Ricinus communis] |