| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
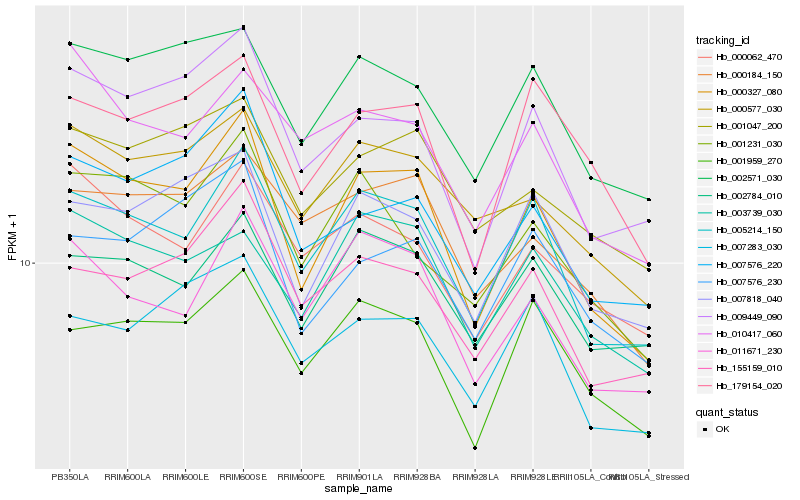
| 1 |
Hb_000327_080 |
0.0 |
- |
- |
PREDICTED: regulator of nonsense transcripts UPF2 [Jatropha curcas] |
| 2 |
Hb_005214_150 |
0.0718159396 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105636019 [Jatropha curcas] |
| 3 |
Hb_009449_090 |
0.0842956157 |
- |
- |
PREDICTED: RNA-binding protein 25 isoform X1 [Jatropha curcas] |
| 4 |
Hb_007818_040 |
0.0911110543 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 50 isoform X2 [Jatropha curcas] |
| 5 |
Hb_011671_230 |
0.0923817521 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002571_030 |
0.0951060498 |
- |
- |
PREDICTED: microfibrillar-associated protein 1-like [Prunus mume] |
| 7 |
Hb_007576_220 |
0.0977215303 |
- |
- |
PREDICTED: kanadaptin [Jatropha curcas] |
| 8 |
Hb_001231_030 |
0.0985302557 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER-like [Jatropha curcas] |
| 9 |
Hb_000577_030 |
0.0990234425 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase 12 -like protein [Gossypium arboreum] |
| 10 |
Hb_001959_270 |
0.0992530581 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH1, mitochondrial isoform X1 [Jatropha curcas] |
| 11 |
Hb_003739_030 |
0.1022335762 |
- |
- |
PREDICTED: putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 [Jatropha curcas] |
| 12 |
Hb_155159_010 |
0.102274128 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 1 [Jatropha curcas] |
| 13 |
Hb_000062_470 |
0.1024518192 |
- |
- |
PREDICTED: cyclin-dependent kinase G-2 [Jatropha curcas] |
| 14 |
Hb_179154_020 |
0.1025234041 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 15 |
Hb_007576_230 |
0.1030439774 |
- |
- |
hypothetical protein JCGZ_25616 [Jatropha curcas] |
| 16 |
Hb_001047_200 |
0.1038526202 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 18 [Eucalyptus grandis] |
| 17 |
Hb_000184_150 |
0.1057914298 |
- |
- |
PREDICTED: U-box domain-containing protein 15 [Jatropha curcas] |
| 18 |
Hb_007283_030 |
0.1060967715 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40C-like [Malus domestica] |
| 19 |
Hb_010417_060 |
0.1069802669 |
- |
- |
PREDICTED: protein SGT1 homolog A-like [Jatropha curcas] |
| 20 |
Hb_002784_010 |
0.107005799 |
- |
- |
DNA-directed RNA polymerase I subunit, putative [Ricinus communis] |