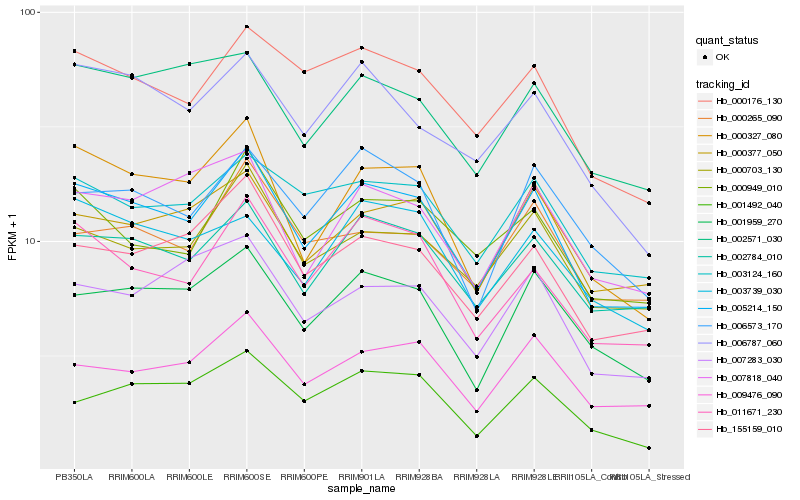
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005214_150 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105636019 [Jatropha curcas] |
| 2 |
Hb_002784_010 |
0.0639990417 |
- |
- |
DNA-directed RNA polymerase I subunit, putative [Ricinus communis] |
| 3 |
Hb_011671_230 |
0.0696938944 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001959_270 |
0.071106571 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH1, mitochondrial isoform X1 [Jatropha curcas] |
| 5 |
Hb_000327_080 |
0.0718159396 |
- |
- |
PREDICTED: regulator of nonsense transcripts UPF2 [Jatropha curcas] |
| 6 |
Hb_000703_130 |
0.0733215061 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 [Jatropha curcas] |
| 7 |
Hb_155159_010 |
0.0740816722 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 1 [Jatropha curcas] |
| 8 |
Hb_000176_130 |
0.0756243151 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Jatropha curcas] |
| 9 |
Hb_009476_090 |
0.0784296765 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 1 [Jatropha curcas] |
| 10 |
Hb_003124_160 |
0.0799546588 |
- |
- |
dynamin, putative [Ricinus communis] |
| 11 |
Hb_000377_050 |
0.0802806434 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 12 |
Hb_000949_010 |
0.0825169721 |
- |
- |
PREDICTED: regulator of nonsense transcripts UPF2 [Jatropha curcas] |
| 13 |
Hb_006573_170 |
0.0848948568 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_007818_040 |
0.0853107664 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 50 isoform X2 [Jatropha curcas] |
| 15 |
Hb_006787_060 |
0.0854124835 |
- |
- |
PREDICTED: probable E3 ubiquitin ligase SUD1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_002571_030 |
0.0854127029 |
- |
- |
PREDICTED: microfibrillar-associated protein 1-like [Prunus mume] |
| 17 |
Hb_007283_030 |
0.0868776471 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40C-like [Malus domestica] |
| 18 |
Hb_000265_090 |
0.0886708727 |
- |
- |
PREDICTED: flowering time control protein FPA [Jatropha curcas] |
| 19 |
Hb_003739_030 |
0.0897808314 |
- |
- |
PREDICTED: putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 [Jatropha curcas] |
| 20 |
Hb_001492_040 |
0.0902647764 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |