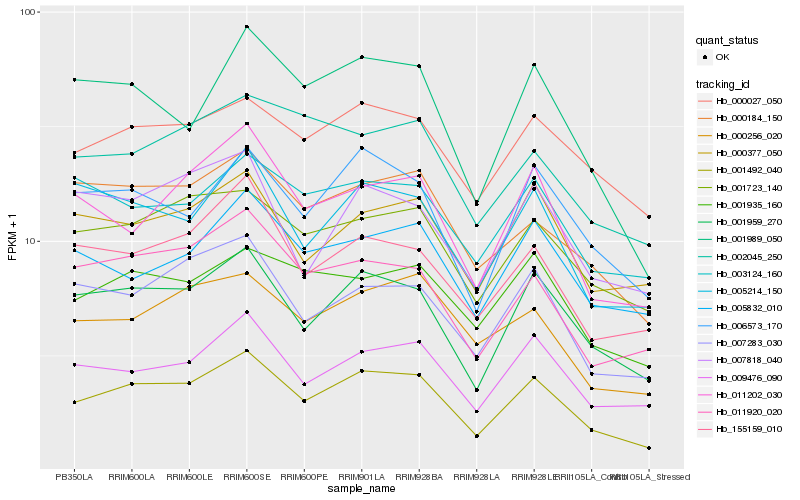
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001492_040 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001959_270 |
0.0628807393 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH1, mitochondrial isoform X1 [Jatropha curcas] |
| 3 |
Hb_006573_170 |
0.0834035337 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001723_140 |
0.0861128684 |
- |
- |
PREDICTED: importin beta-like SAD2 homolog isoform X2 [Jatropha curcas] |
| 5 |
Hb_009476_090 |
0.0864255292 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 1 [Jatropha curcas] |
| 6 |
Hb_001989_050 |
0.0875252316 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 5 [Jatropha curcas] |
| 7 |
Hb_007283_030 |
0.0879774408 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40C-like [Malus domestica] |
| 8 |
Hb_011920_020 |
0.0888711786 |
- |
- |
PREDICTED: uncharacterized protein LOC105645397 [Jatropha curcas] |
| 9 |
Hb_011202_030 |
0.0902265674 |
- |
- |
arsenite-resistance protein, putative [Ricinus communis] |
| 10 |
Hb_005214_150 |
0.0902647764 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105636019 [Jatropha curcas] |
| 11 |
Hb_155159_010 |
0.0907993981 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 1 [Jatropha curcas] |
| 12 |
Hb_000184_150 |
0.0911296476 |
- |
- |
PREDICTED: U-box domain-containing protein 15 [Jatropha curcas] |
| 13 |
Hb_000027_050 |
0.0917318714 |
- |
- |
PREDICTED: uncharacterized protein LOC105636933 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000256_020 |
0.0926444896 |
transcription factor |
TF Family: PHD |
PREDICTED: CHD3-type chromatin-remodeling factor PICKLE isoform X1 [Jatropha curcas] |
| 15 |
Hb_001935_160 |
0.0944185028 |
- |
- |
PREDICTED: F-box protein At5g51370-like [Jatropha curcas] |
| 16 |
Hb_000377_050 |
0.0975803493 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 17 |
Hb_002045_250 |
0.1010911858 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP63 isoform X3 [Jatropha curcas] |
| 18 |
Hb_005832_010 |
0.1012664164 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 19 |
Hb_007818_040 |
0.1033765705 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 50 isoform X2 [Jatropha curcas] |
| 20 |
Hb_003124_160 |
0.1037299028 |
- |
- |
dynamin, putative [Ricinus communis] |