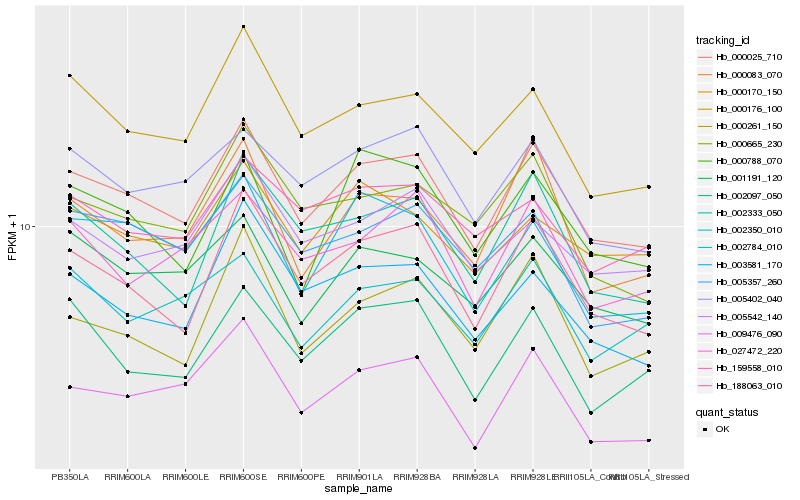
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000025_710 |
0.0 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |
| 2 |
Hb_188063_010 |
0.0486479356 |
- |
- |
hypothetical protein RCOM_1469330 [Ricinus communis] |
| 3 |
Hb_005402_040 |
0.0601328658 |
- |
- |
PREDICTED: replication factor C subunit 1 [Jatropha curcas] |
| 4 |
Hb_001191_120 |
0.0616783509 |
- |
- |
PREDICTED: cleavage stimulating factor 64 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000176_100 |
0.0618062793 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Solanum lycopersicum] |
| 6 |
Hb_002333_050 |
0.0633750283 |
- |
- |
PREDICTED: uncharacterized protein LOC105644854 isoform X1 [Jatropha curcas] |
| 7 |
Hb_005542_140 |
0.0650181566 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 8 |
Hb_000083_070 |
0.0659071379 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 1 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000170_150 |
0.0664846252 |
- |
- |
PREDICTED: uncharacterized protein LOC105638996 isoform X3 [Jatropha curcas] |
| 10 |
Hb_002784_010 |
0.0680971001 |
- |
- |
DNA-directed RNA polymerase I subunit, putative [Ricinus communis] |
| 11 |
Hb_027472_220 |
0.0686425775 |
- |
- |
PREDICTED: nucleolar complex protein 3 homolog isoform X1 [Jatropha curcas] |
| 12 |
Hb_005357_260 |
0.0688601151 |
- |
- |
PREDICTED: polyadenylation and cleavage factor homolog 4 [Jatropha curcas] |
| 13 |
Hb_000261_150 |
0.0699757882 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003581_170 |
0.0699872024 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 15 |
Hb_002350_010 |
0.0700866628 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit rpc1 [Jatropha curcas] |
| 16 |
Hb_000665_230 |
0.0703042478 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 17 |
Hb_009476_090 |
0.0706543742 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 1 [Jatropha curcas] |
| 18 |
Hb_159558_010 |
0.0715851124 |
- |
- |
PREDICTED: uncharacterized protein LOC105635846 [Jatropha curcas] |
| 19 |
Hb_000788_070 |
0.0718215505 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g10270 [Jatropha curcas] |
| 20 |
Hb_002097_050 |
0.0722262599 |
- |
- |
PREDICTED: protein fluG isoform X1 [Jatropha curcas] |