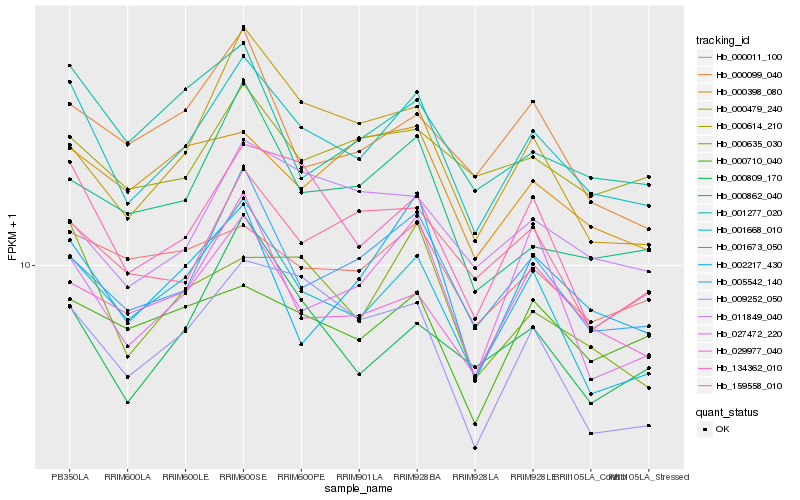
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001668_010 |
0.0 |
- |
- |
PREDICTED: RNA-binding protein with multiple splicing [Jatropha curcas] |
| 2 |
Hb_001673_050 |
0.0554646703 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_134362_010 |
0.0597116329 |
- |
- |
PREDICTED: FAS-associated factor 2 [Jatropha curcas] |
| 4 |
Hb_005542_140 |
0.0617038134 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 5 |
Hb_000635_030 |
0.0635285892 |
- |
- |
mannosyl-oligosaccharide glucosidase, putative [Ricinus communis] |
| 6 |
Hb_027472_220 |
0.0733572206 |
- |
- |
PREDICTED: nucleolar complex protein 3 homolog isoform X1 [Jatropha curcas] |
| 7 |
Hb_000011_100 |
0.0740817731 |
- |
- |
PREDICTED: uncharacterized protein C630.12 [Jatropha curcas] |
| 8 |
Hb_029977_040 |
0.0744496196 |
- |
- |
PREDICTED: uncharacterized protein LOC105640155 isoform X4 [Jatropha curcas] |
| 9 |
Hb_001277_020 |
0.075113812 |
- |
- |
PREDICTED: armadillo repeat-containing protein LFR [Jatropha curcas] |
| 10 |
Hb_009252_050 |
0.075900359 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 11 |
Hb_159558_010 |
0.0759090472 |
- |
- |
PREDICTED: uncharacterized protein LOC105635846 [Jatropha curcas] |
| 12 |
Hb_000862_040 |
0.0759872597 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 13 |
Hb_000710_040 |
0.0761582844 |
- |
- |
PREDICTED: HEAT repeat-containing protein 6 [Jatropha curcas] |
| 14 |
Hb_002217_430 |
0.0769318502 |
- |
- |
PREDICTED: flowering time control protein FCA isoform X4 [Jatropha curcas] |
| 15 |
Hb_000809_170 |
0.0772690056 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit beta-like protein 1 homolog isoform X1 [Jatropha curcas] |
| 16 |
Hb_000614_210 |
0.0784741745 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_011849_040 |
0.0787316943 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 18 |
Hb_000398_080 |
0.0788692434 |
- |
- |
tip120, putative [Ricinus communis] |
| 19 |
Hb_000099_040 |
0.078936262 |
- |
- |
unnamed protein product [Coffea canephora] |
| 20 |
Hb_000479_240 |
0.0790231688 |
- |
- |
PREDICTED: probable ubiquitin conjugation factor E4 isoform X2 [Jatropha curcas] |