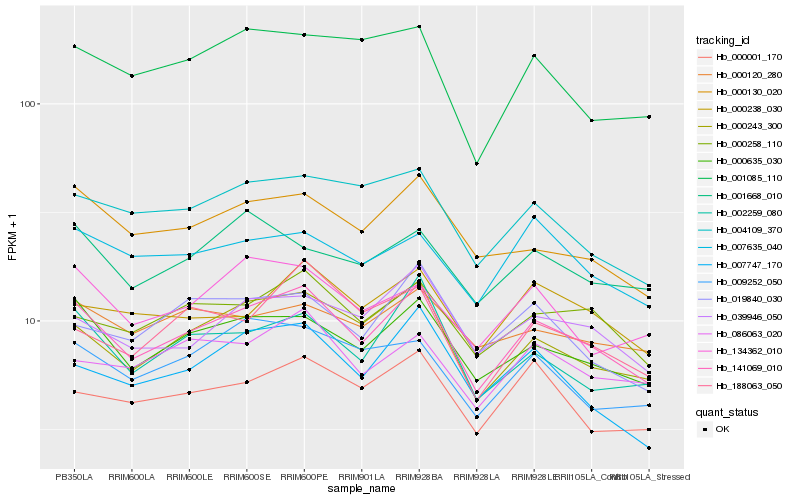
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_141069_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000635_030 |
0.0593698138 |
- |
- |
mannosyl-oligosaccharide glucosidase, putative [Ricinus communis] |
| 3 |
Hb_000243_300 |
0.0615399864 |
- |
- |
catalytic, putative [Ricinus communis] |
| 4 |
Hb_000258_110 |
0.0762198484 |
- |
- |
PREDICTED: probable apyrase 6 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000130_020 |
0.0783214502 |
- |
- |
Phosphoenolpyruvate carboxylase, putative [Ricinus communis] |
| 6 |
Hb_002259_080 |
0.0786137951 |
- |
- |
protein transport protein sec23, putative [Ricinus communis] |
| 7 |
Hb_039946_050 |
0.0797885306 |
- |
- |
catalytic, putative [Ricinus communis] |
| 8 |
Hb_004109_370 |
0.0831308858 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 9 |
Hb_000238_030 |
0.0841229214 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_000001_170 |
0.0850767904 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 11 |
Hb_086063_020 |
0.0854408513 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 [Jatropha curcas] |
| 12 |
Hb_007747_170 |
0.0867014292 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |
| 13 |
Hb_009252_050 |
0.0878334552 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 14 |
Hb_134362_010 |
0.0881580396 |
- |
- |
PREDICTED: FAS-associated factor 2 [Jatropha curcas] |
| 15 |
Hb_007635_040 |
0.0886342652 |
- |
- |
PREDICTED: WAT1-related protein At3g45870 isoform X1 [Jatropha curcas] |
| 16 |
Hb_188063_050 |
0.0893178489 |
- |
- |
PREDICTED: putative G3BP-like protein [Jatropha curcas] |
| 17 |
Hb_001668_010 |
0.089739439 |
- |
- |
PREDICTED: RNA-binding protein with multiple splicing [Jatropha curcas] |
| 18 |
Hb_001085_110 |
0.0899496431 |
- |
- |
T6D22.2 [Arabidopsis thaliana] |
| 19 |
Hb_019840_030 |
0.0907385868 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000120_280 |
0.0921057501 |
- |
- |
PREDICTED: protein transport protein SEC23 [Jatropha curcas] |