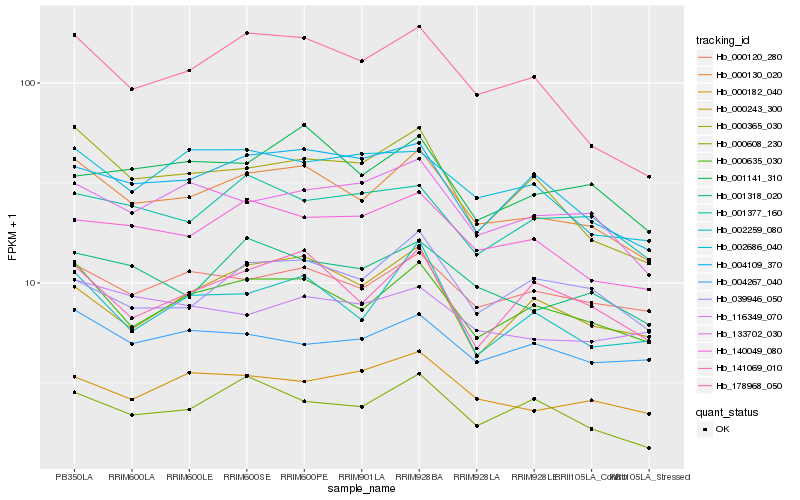
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000130_020 |
0.0 |
- |
- |
Phosphoenolpyruvate carboxylase, putative [Ricinus communis] |
| 2 |
Hb_000635_030 |
0.0519722529 |
- |
- |
mannosyl-oligosaccharide glucosidase, putative [Ricinus communis] |
| 3 |
Hb_178968_050 |
0.0718968239 |
- |
- |
PREDICTED: probable serine incorporator [Jatropha curcas] |
| 4 |
Hb_000120_280 |
0.0742270171 |
- |
- |
PREDICTED: protein transport protein SEC23 [Jatropha curcas] |
| 5 |
Hb_140049_080 |
0.0746130247 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Jatropha curcas] |
| 6 |
Hb_001318_020 |
0.0754810259 |
- |
- |
HAT dimerization domain-containing protein isoform 2 [Theobroma cacao] |
| 7 |
Hb_141069_010 |
0.0783214502 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000365_030 |
0.078921053 |
- |
- |
PREDICTED: transmembrane protein 87B [Jatropha curcas] |
| 9 |
Hb_004109_370 |
0.0793742511 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 10 |
Hb_002259_080 |
0.0814143602 |
- |
- |
protein transport protein sec23, putative [Ricinus communis] |
| 11 |
Hb_133702_030 |
0.0821231174 |
- |
- |
PREDICTED: uric acid degradation bifunctional protein TTL isoform X1 [Jatropha curcas] |
| 12 |
Hb_000608_230 |
0.0831149047 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 13 |
Hb_000182_040 |
0.0879160856 |
transcription factor |
TF Family: SNF2 |
PREDICTED: protein CHROMATIN REMODELING 24 [Jatropha curcas] |
| 14 |
Hb_002686_040 |
0.0898187516 |
- |
- |
PREDICTED: T-complex protein 1 subunit theta [Jatropha curcas] |
| 15 |
Hb_001141_310 |
0.0903230275 |
- |
- |
PREDICTED: uncharacterized protein LOC105632212 [Jatropha curcas] |
| 16 |
Hb_004267_040 |
0.0910690803 |
- |
- |
PREDICTED: transcriptional adapter ADA2a [Jatropha curcas] |
| 17 |
Hb_039946_050 |
0.0910810585 |
- |
- |
catalytic, putative [Ricinus communis] |
| 18 |
Hb_116349_070 |
0.0912293151 |
- |
- |
hypothetical protein JCGZ_21753 [Jatropha curcas] |
| 19 |
Hb_001377_160 |
0.0916266477 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 20 |
Hb_000243_300 |
0.0919654335 |
- |
- |
catalytic, putative [Ricinus communis] |