| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
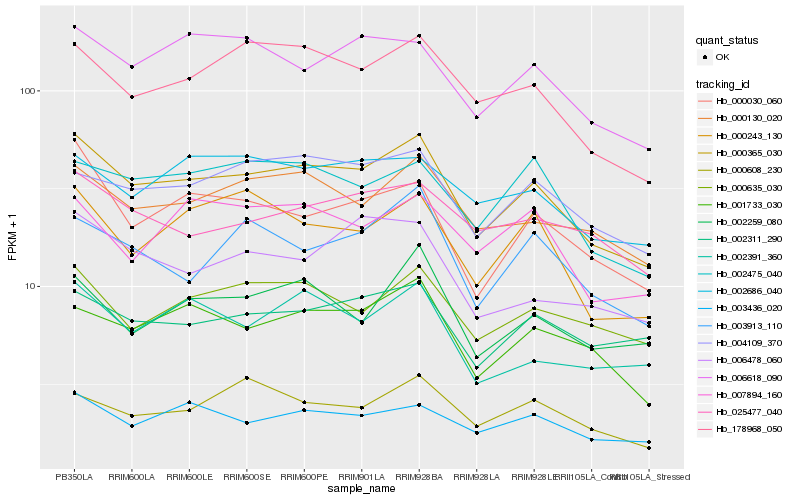
Hb_000365_030 |
0.0 |
- |
- |
PREDICTED: transmembrane protein 87B [Jatropha curcas] |
| 2 |
Hb_000130_020 |
0.078921053 |
- |
- |
Phosphoenolpyruvate carboxylase, putative [Ricinus communis] |
| 3 |
Hb_178968_050 |
0.0790924433 |
- |
- |
PREDICTED: probable serine incorporator [Jatropha curcas] |
| 4 |
Hb_001733_030 |
0.0807510244 |
- |
- |
PREDICTED: uncharacterized protein LOC100261386 isoform X2 [Vitis vinifera] |
| 5 |
Hb_004109_370 |
0.0837446167 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 6 |
Hb_000030_060 |
0.0871155145 |
- |
- |
PREDICTED: T-complex protein 1 subunit theta [Jatropha curcas] |
| 7 |
Hb_000635_030 |
0.088533042 |
- |
- |
mannosyl-oligosaccharide glucosidase, putative [Ricinus communis] |
| 8 |
Hb_003436_020 |
0.0888245455 |
transcription factor |
TF Family: Orphans |
PREDICTED: uncharacterized protein LOC105636276 isoform X2 [Jatropha curcas] |
| 9 |
Hb_006618_090 |
0.0888728494 |
- |
- |
PREDICTED: FAM10 family protein At4g22670 [Jatropha curcas] |
| 10 |
Hb_002475_040 |
0.0903617337 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 11 |
Hb_000243_130 |
0.0921396358 |
- |
- |
hypothetical protein M569_10795, partial [Genlisea aurea] |
| 12 |
Hb_003913_110 |
0.0923417441 |
- |
- |
PREDICTED: uncharacterized protein LOC105640319 [Jatropha curcas] |
| 13 |
Hb_002311_290 |
0.0923699732 |
- |
- |
PREDICTED: pumilio homolog 24 [Jatropha curcas] |
| 14 |
Hb_002686_040 |
0.0938592644 |
- |
- |
PREDICTED: T-complex protein 1 subunit theta [Jatropha curcas] |
| 15 |
Hb_025477_040 |
0.0940930954 |
- |
- |
PREDICTED: post-GPI attachment to proteins factor 3 [Jatropha curcas] |
| 16 |
Hb_002259_080 |
0.0962668195 |
- |
- |
protein transport protein sec23, putative [Ricinus communis] |
| 17 |
Hb_000608_230 |
0.0968012212 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 18 |
Hb_002391_360 |
0.0970606301 |
- |
- |
hypothetical protein CICLE_v10026208mg [Citrus clementina] |
| 19 |
Hb_007894_160 |
0.0971287034 |
- |
- |
PREDICTED: purple acid phosphatase 18 [Jatropha curcas] |
| 20 |
Hb_006478_060 |
0.0972385413 |
- |
- |
PREDICTED: homocysteine S-methyltransferase 2 isoform X1 [Jatropha curcas] |