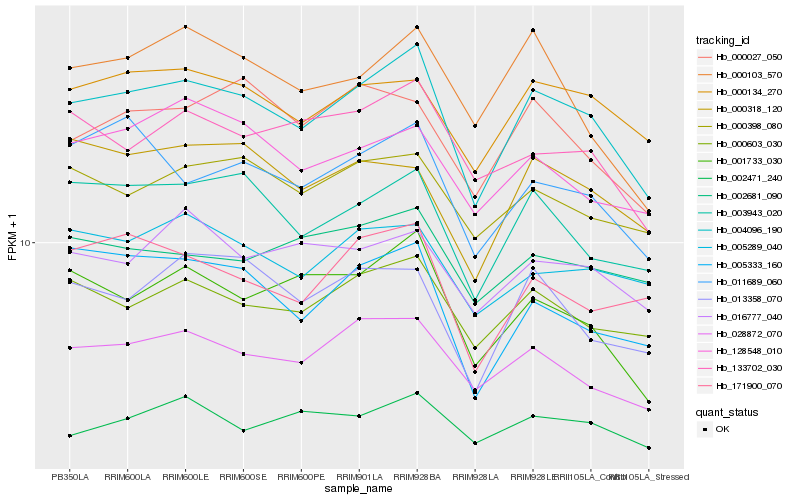
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004096_190 |
0.0 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 2 |
Hb_028872_070 |
0.072279588 |
desease resistance |
Gene Name: DEAD |
PREDICTED: putative pre-mRNA-splicing factor ATP-dependent RNA helicase PRP1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000603_030 |
0.0759635886 |
- |
- |
Ribonuclease III, putative [Ricinus communis] |
| 4 |
Hb_003943_020 |
0.0762048884 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase PASTICCINO1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000134_270 |
0.0793178739 |
- |
- |
arginine/serine rich splicing factor sf4/14, putative [Ricinus communis] |
| 6 |
Hb_001733_030 |
0.0854500448 |
- |
- |
PREDICTED: uncharacterized protein LOC100261386 isoform X2 [Vitis vinifera] |
| 7 |
Hb_000398_080 |
0.0868475674 |
- |
- |
tip120, putative [Ricinus communis] |
| 8 |
Hb_011689_060 |
0.0874364586 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RF298 [Jatropha curcas] |
| 9 |
Hb_133702_030 |
0.0876755489 |
- |
- |
PREDICTED: uric acid degradation bifunctional protein TTL isoform X1 [Jatropha curcas] |
| 10 |
Hb_005289_040 |
0.0909606988 |
- |
- |
poly(A) polymerase, putative [Ricinus communis] |
| 11 |
Hb_000027_050 |
0.0911931245 |
- |
- |
PREDICTED: uncharacterized protein LOC105636933 isoform X1 [Jatropha curcas] |
| 12 |
Hb_013358_070 |
0.0916123265 |
- |
- |
protein with unknown function [Ricinus communis] |
| 13 |
Hb_016777_040 |
0.0920477807 |
- |
- |
PREDICTED: DNA polymerase eta-like [Jatropha curcas] |
| 14 |
Hb_005333_160 |
0.092081445 |
- |
- |
hypothetical protein B456_009G230200 [Gossypium raimondii] |
| 15 |
Hb_171900_070 |
0.0924893028 |
- |
- |
- |
| 16 |
Hb_002471_240 |
0.0934497713 |
- |
- |
PREDICTED: protein POLLEN DEFECTIVE IN GUIDANCE 1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000103_570 |
0.0934554779 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic [Jatropha curcas] |
| 18 |
Hb_128548_010 |
0.0935216899 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000318_120 |
0.0937873216 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: DNA ligase 1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_002681_090 |
0.0948529135 |
- |
- |
PREDICTED: golgin candidate 6 isoform X1 [Jatropha curcas] |