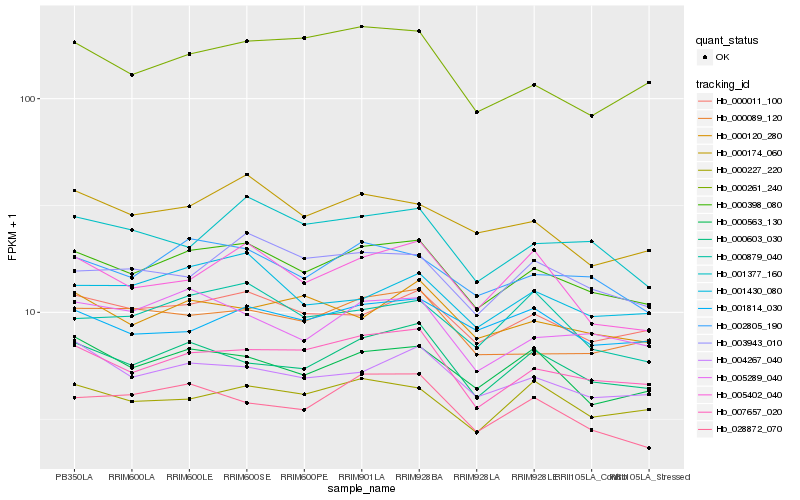
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000398_080 |
0.0 |
- |
- |
tip120, putative [Ricinus communis] |
| 2 |
Hb_007657_020 |
0.0461781073 |
- |
- |
PREDICTED: PRA1 family protein H isoform X1 [Jatropha curcas] |
| 3 |
Hb_002805_190 |
0.0469328932 |
- |
- |
spliceosome associated protein, putative [Ricinus communis] |
| 4 |
Hb_000011_100 |
0.0502174685 |
- |
- |
PREDICTED: uncharacterized protein C630.12 [Jatropha curcas] |
| 5 |
Hb_000603_030 |
0.0528627955 |
- |
- |
Ribonuclease III, putative [Ricinus communis] |
| 6 |
Hb_004267_040 |
0.0586951249 |
- |
- |
PREDICTED: transcriptional adapter ADA2a [Jatropha curcas] |
| 7 |
Hb_000563_130 |
0.0591844314 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 8 |
Hb_000174_060 |
0.0605706099 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 9 |
Hb_005289_040 |
0.0616782815 |
- |
- |
poly(A) polymerase, putative [Ricinus communis] |
| 10 |
Hb_000227_220 |
0.0617134287 |
- |
- |
PREDICTED: protein phosphatase 2C 70 [Jatropha curcas] |
| 11 |
Hb_003943_010 |
0.0627797563 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001430_080 |
0.063217617 |
- |
- |
PREDICTED: putative mediator of RNA polymerase II transcription subunit 26 [Jatropha curcas] |
| 13 |
Hb_001377_160 |
0.0647237812 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 14 |
Hb_000879_040 |
0.065412086 |
- |
- |
PREDICTED: calcium-transporting ATPase 3, endoplasmic reticulum-type [Jatropha curcas] |
| 15 |
Hb_028872_070 |
0.0662134761 |
desease resistance |
Gene Name: DEAD |
PREDICTED: putative pre-mRNA-splicing factor ATP-dependent RNA helicase PRP1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000089_120 |
0.0663620833 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 17 |
Hb_005402_040 |
0.0679172693 |
- |
- |
PREDICTED: replication factor C subunit 1 [Jatropha curcas] |
| 18 |
Hb_000120_280 |
0.0680866473 |
- |
- |
PREDICTED: protein transport protein SEC23 [Jatropha curcas] |
| 19 |
Hb_001814_030 |
0.0687444325 |
- |
- |
PREDICTED: uncharacterized protein LOC105650634 [Jatropha curcas] |
| 20 |
Hb_000261_240 |
0.0695489896 |
- |
- |
RecName: Full=Elongation factor 1-alpha; Short=EF-1-alpha [Manihot esculenta] |