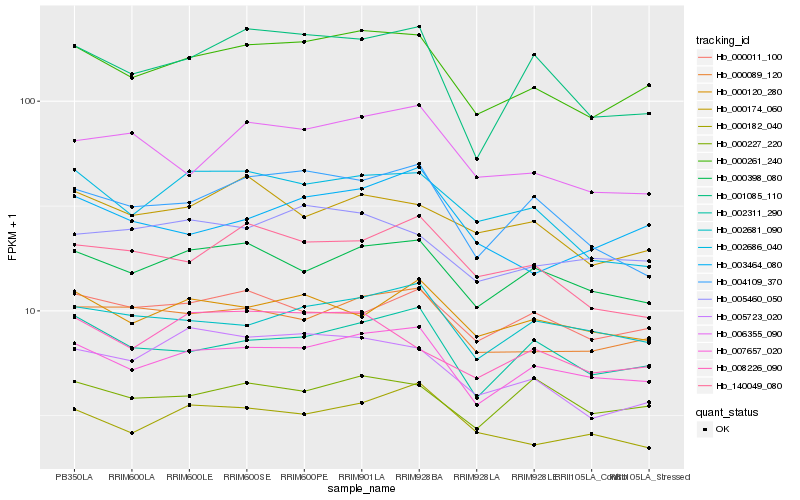
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000261_240 |
0.0 |
- |
- |
RecName: Full=Elongation factor 1-alpha; Short=EF-1-alpha [Manihot esculenta] |
| 2 |
Hb_007657_020 |
0.0474741017 |
- |
- |
PREDICTED: PRA1 family protein H isoform X1 [Jatropha curcas] |
| 3 |
Hb_000089_120 |
0.0622459978 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 4 |
Hb_002311_290 |
0.0692523297 |
- |
- |
PREDICTED: pumilio homolog 24 [Jatropha curcas] |
| 5 |
Hb_000398_080 |
0.0695489896 |
- |
- |
tip120, putative [Ricinus communis] |
| 6 |
Hb_005460_050 |
0.0749050264 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 7 |
Hb_005723_020 |
0.0754201961 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001085_110 |
0.0767734741 |
- |
- |
T6D22.2 [Arabidopsis thaliana] |
| 9 |
Hb_002681_090 |
0.0776700552 |
- |
- |
PREDICTED: golgin candidate 6 isoform X1 [Jatropha curcas] |
| 10 |
Hb_008226_090 |
0.0777178994 |
- |
- |
PREDICTED: reticulon-like protein B16 isoform X1 [Jatropha curcas] |
| 11 |
Hb_006355_090 |
0.0789090146 |
desease resistance |
Gene Name: AAA |
PREDICTED: 26S proteasome regulatory subunit 4 homolog A [Jatropha curcas] |
| 12 |
Hb_002686_040 |
0.078967538 |
- |
- |
PREDICTED: T-complex protein 1 subunit theta [Jatropha curcas] |
| 13 |
Hb_000174_060 |
0.0796321371 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 14 |
Hb_000182_040 |
0.0805110343 |
transcription factor |
TF Family: SNF2 |
PREDICTED: protein CHROMATIN REMODELING 24 [Jatropha curcas] |
| 15 |
Hb_004109_370 |
0.0809025842 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 16 |
Hb_000011_100 |
0.0820479529 |
- |
- |
PREDICTED: uncharacterized protein C630.12 [Jatropha curcas] |
| 17 |
Hb_000120_280 |
0.0827661255 |
- |
- |
PREDICTED: protein transport protein SEC23 [Jatropha curcas] |
| 18 |
Hb_000227_220 |
0.0830384092 |
- |
- |
PREDICTED: protein phosphatase 2C 70 [Jatropha curcas] |
| 19 |
Hb_003464_080 |
0.0832287399 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme-like [Jatropha curcas] |
| 20 |
Hb_140049_080 |
0.0835577642 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Jatropha curcas] |