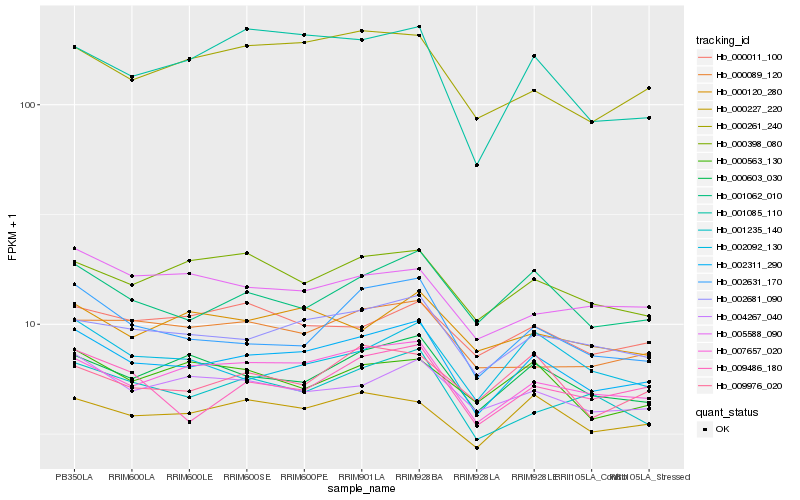
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002311_290 |
0.0 |
- |
- |
PREDICTED: pumilio homolog 24 [Jatropha curcas] |
| 2 |
Hb_007657_020 |
0.0488065064 |
- |
- |
PREDICTED: PRA1 family protein H isoform X1 [Jatropha curcas] |
| 3 |
Hb_002681_090 |
0.0522075178 |
- |
- |
PREDICTED: golgin candidate 6 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000603_030 |
0.0614154052 |
- |
- |
Ribonuclease III, putative [Ricinus communis] |
| 5 |
Hb_001062_010 |
0.0636671781 |
- |
- |
PREDICTED: CWF19-like protein 2 [Jatropha curcas] |
| 6 |
Hb_002631_170 |
0.0645424246 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 36 [Jatropha curcas] |
| 7 |
Hb_000227_220 |
0.0672715224 |
- |
- |
PREDICTED: protein phosphatase 2C 70 [Jatropha curcas] |
| 8 |
Hb_004267_040 |
0.0683546779 |
- |
- |
PREDICTED: transcriptional adapter ADA2a [Jatropha curcas] |
| 9 |
Hb_000261_240 |
0.0692523297 |
- |
- |
RecName: Full=Elongation factor 1-alpha; Short=EF-1-alpha [Manihot esculenta] |
| 10 |
Hb_001235_140 |
0.0694215198 |
- |
- |
PREDICTED: protein arginine N-methyltransferase 1.6 [Jatropha curcas] |
| 11 |
Hb_009486_180 |
0.0697817279 |
- |
- |
unknown [Glycine max] |
| 12 |
Hb_000398_080 |
0.0709579389 |
- |
- |
tip120, putative [Ricinus communis] |
| 13 |
Hb_002092_130 |
0.0742050444 |
- |
- |
PREDICTED: protein CASP-like [Oryza brachyantha] |
| 14 |
Hb_009976_020 |
0.0771002695 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105631286 [Jatropha curcas] |
| 15 |
Hb_000011_100 |
0.0773424501 |
- |
- |
PREDICTED: uncharacterized protein C630.12 [Jatropha curcas] |
| 16 |
Hb_001085_110 |
0.0782156105 |
- |
- |
T6D22.2 [Arabidopsis thaliana] |
| 17 |
Hb_000563_130 |
0.0782789287 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 18 |
Hb_000089_120 |
0.0784385284 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 19 |
Hb_000120_280 |
0.079130686 |
- |
- |
PREDICTED: protein transport protein SEC23 [Jatropha curcas] |
| 20 |
Hb_005588_090 |
0.0791681036 |
- |
- |
hypothetical protein CICLE_v10003480mg [Citrus clementina] |