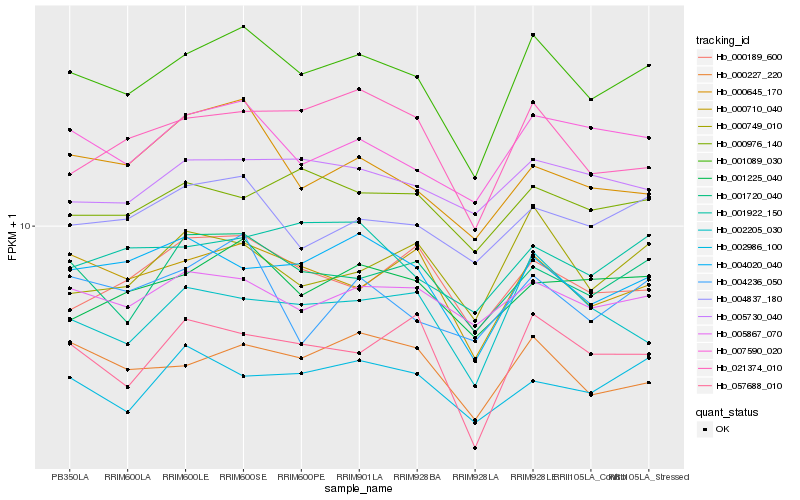
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001089_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638026 [Jatropha curcas] |
| 2 |
Hb_000710_040 |
0.0570552817 |
- |
- |
PREDICTED: HEAT repeat-containing protein 6 [Jatropha curcas] |
| 3 |
Hb_007590_020 |
0.0584005894 |
- |
- |
PREDICTED: protein DAMAGED DNA-BINDING 2 [Jatropha curcas] |
| 4 |
Hb_000227_220 |
0.0609112783 |
- |
- |
PREDICTED: protein phosphatase 2C 70 [Jatropha curcas] |
| 5 |
Hb_001720_040 |
0.0610647007 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004020_040 |
0.0647039998 |
- |
- |
PREDICTED: GTP-binding protein At3g49725, chloroplastic [Jatropha curcas] |
| 7 |
Hb_004837_180 |
0.0647549213 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_005867_070 |
0.0654802044 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_001225_040 |
0.0660039446 |
- |
- |
PREDICTED: meiosis-specific nuclear structural protein 1 isoform X5 [Jatropha curcas] |
| 10 |
Hb_057688_010 |
0.0693435528 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000749_010 |
0.0700562639 |
- |
- |
PREDICTED: non-canonical poly(A) RNA polymerase PAPD5 isoform X1 [Jatropha curcas] |
| 12 |
Hb_021374_010 |
0.0701892071 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-A1 [Populus euphratica] |
| 13 |
Hb_001922_150 |
0.0705276339 |
- |
- |
PREDICTED: WD repeat-containing protein 91 homolog isoform X1 [Jatropha curcas] |
| 14 |
Hb_002205_030 |
0.0712608884 |
- |
- |
PREDICTED: alpha-galactosidase 3 [Jatropha curcas] |
| 15 |
Hb_000976_140 |
0.07153882 |
- |
- |
PREDICTED: uncharacterized protein LOC105646208 isoform X1 [Jatropha curcas] |
| 16 |
Hb_005730_040 |
0.0718555047 |
- |
- |
hypothetical protein PRUPE_ppa004338mg [Prunus persica] |
| 17 |
Hb_004236_050 |
0.0736311454 |
- |
- |
PREDICTED: nipped-B-like protein A [Jatropha curcas] |
| 18 |
Hb_000645_170 |
0.0742523703 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002986_100 |
0.0743414675 |
- |
- |
PREDICTED: uncharacterized protein LOC105639686 [Jatropha curcas] |
| 20 |
Hb_000189_600 |
0.0745035245 |
- |
- |
PREDICTED: protein MON2 homolog isoform X1 [Jatropha curcas] |