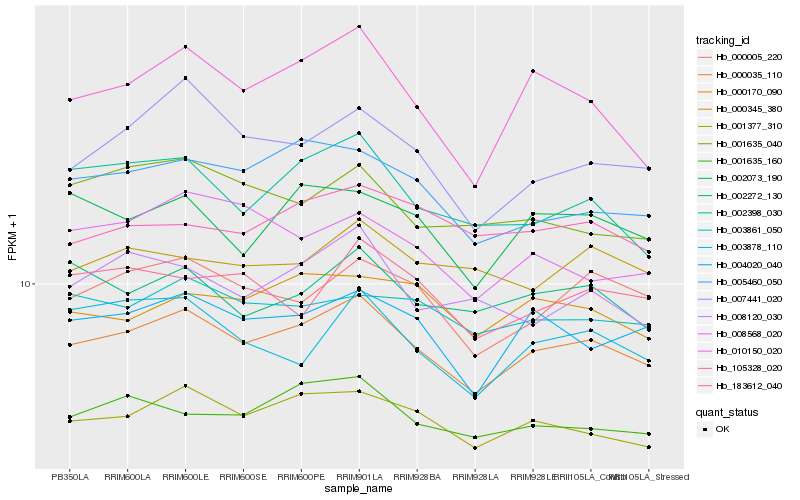
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000035_110 |
0.0 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 5 [Jatropha curcas] |
| 2 |
Hb_001377_310 |
0.0474938008 |
transcription factor |
TF Family: Coactivator p15 |
PREDICTED: uncharacterized protein LOC105642839 [Jatropha curcas] |
| 3 |
Hb_000005_220 |
0.0589235925 |
- |
- |
DNA ligase 1, partial [Glycine soja] |
| 4 |
Hb_004020_040 |
0.0593991075 |
- |
- |
PREDICTED: GTP-binding protein At3g49725, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001635_160 |
0.0610885774 |
- |
- |
hypothetical protein JCGZ_24810 [Jatropha curcas] |
| 6 |
Hb_005460_050 |
0.0615553452 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 7 |
Hb_010150_020 |
0.06414981 |
transcription factor |
TF Family: HMG |
PREDICTED: FACT complex subunit SSRP1 [Jatropha curcas] |
| 8 |
Hb_002398_030 |
0.0647491226 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 9 |
Hb_007441_020 |
0.0648206015 |
- |
- |
Rab6 [Hevea brasiliensis] |
| 10 |
Hb_002073_190 |
0.0667802134 |
- |
- |
PREDICTED: uncharacterized protein LOC105649812 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003861_050 |
0.0676109396 |
- |
- |
PREDICTED: uncharacterized protein LOC105650779 [Jatropha curcas] |
| 12 |
Hb_002272_130 |
0.0682448687 |
- |
- |
PREDICTED: DNA excision repair protein ERCC-1 [Jatropha curcas] |
| 13 |
Hb_000170_090 |
0.0693542249 |
- |
- |
PREDICTED: UDP-sugar pyrophosphorylase [Jatropha curcas] |
| 14 |
Hb_008120_030 |
0.0707927424 |
- |
- |
PREDICTED: uncharacterized protein LOC101216675 [Cucumis sativus] |
| 15 |
Hb_183612_040 |
0.0735638909 |
- |
- |
PREDICTED: uncharacterized protein LOC105632370 [Jatropha curcas] |
| 16 |
Hb_105328_020 |
0.0736387501 |
- |
- |
PREDICTED: phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN [Jatropha curcas] |
| 17 |
Hb_000345_380 |
0.073824095 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 7, peroxisomal [Jatropha curcas] |
| 18 |
Hb_001635_040 |
0.0739875349 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Gossypium raimondii] |
| 19 |
Hb_003878_110 |
0.0746645263 |
- |
- |
PREDICTED: uncharacterized protein LOC105637024 [Jatropha curcas] |
| 20 |
Hb_008568_020 |
0.0749257654 |
- |
- |
PREDICTED: uncharacterized protein LOC105122325 isoform X3 [Populus euphratica] |