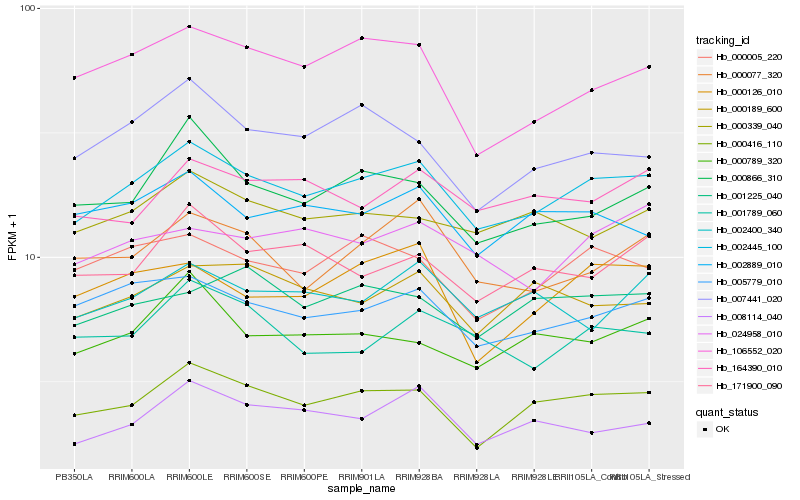
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002445_100 |
0.0 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 5 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000416_110 |
0.0565281346 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RFK1 isoform X2 [Jatropha curcas] |
| 3 |
Hb_005779_010 |
0.0630142704 |
- |
- |
catalytic, putative [Ricinus communis] |
| 4 |
Hb_000005_220 |
0.066637458 |
- |
- |
DNA ligase 1, partial [Glycine soja] |
| 5 |
Hb_000339_040 |
0.0735155977 |
- |
- |
PREDICTED: SAP30-binding protein isoform X2 [Jatropha curcas] |
| 6 |
Hb_106552_020 |
0.075052124 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor RS2Z33 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002889_010 |
0.0750899579 |
- |
- |
ubx domain-containing, putative [Ricinus communis] |
| 8 |
Hb_000866_310 |
0.0757218304 |
- |
- |
PREDICTED: neural Wiskott-Aldrich syndrome protein [Jatropha curcas] |
| 9 |
Hb_164390_010 |
0.0768388155 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1a [Jatropha curcas] |
| 10 |
Hb_000077_320 |
0.0777714508 |
- |
- |
PREDICTED: lanC-like protein GCL2 [Jatropha curcas] |
| 11 |
Hb_000189_600 |
0.0778333761 |
- |
- |
PREDICTED: protein MON2 homolog isoform X1 [Jatropha curcas] |
| 12 |
Hb_171900_090 |
0.0778391442 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001225_040 |
0.078055381 |
- |
- |
PREDICTED: meiosis-specific nuclear structural protein 1 isoform X5 [Jatropha curcas] |
| 14 |
Hb_000126_010 |
0.0781016163 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RFK1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000789_320 |
0.0782184013 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_008114_040 |
0.0786327791 |
- |
- |
Fanconi anemia group D2 [Gossypium arboreum] |
| 17 |
Hb_002400_340 |
0.0799626518 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 10 [Jatropha curcas] |
| 18 |
Hb_024958_010 |
0.0799916493 |
- |
- |
PREDICTED: LRR repeats and ubiquitin-like domain-containing protein At2g30105 [Jatropha curcas] |
| 19 |
Hb_007441_020 |
0.0809580345 |
- |
- |
Rab6 [Hevea brasiliensis] |
| 20 |
Hb_001789_060 |
0.0815927098 |
- |
- |
calmodulin-binding heat-shock protein, putative [Ricinus communis] |