| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
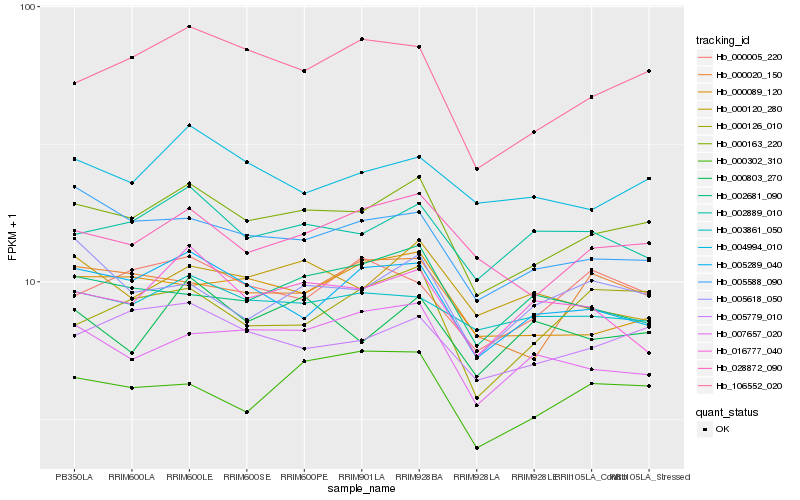
Hb_000163_220 |
0.0 |
- |
- |
hypothetical protein CISIN_1g022301mg [Citrus sinensis] |
| 2 |
Hb_106552_020 |
0.0618830741 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor RS2Z33 isoform X1 [Jatropha curcas] |
| 3 |
Hb_005779_010 |
0.0630555432 |
- |
- |
catalytic, putative [Ricinus communis] |
| 4 |
Hb_028872_090 |
0.0641460514 |
- |
- |
PREDICTED: urease [Jatropha curcas] |
| 5 |
Hb_005588_090 |
0.0667787662 |
- |
- |
hypothetical protein CICLE_v10003480mg [Citrus clementina] |
| 6 |
Hb_005289_040 |
0.0678624764 |
- |
- |
poly(A) polymerase, putative [Ricinus communis] |
| 7 |
Hb_002889_010 |
0.0708711893 |
- |
- |
ubx domain-containing, putative [Ricinus communis] |
| 8 |
Hb_000089_120 |
0.0722240267 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 9 |
Hb_000020_150 |
0.0733336893 |
- |
- |
PREDICTED: uncharacterized protein LOC105636325 [Jatropha curcas] |
| 10 |
Hb_003861_050 |
0.0740789696 |
- |
- |
PREDICTED: uncharacterized protein LOC105650779 [Jatropha curcas] |
| 11 |
Hb_004994_010 |
0.0748531209 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 1 [Jatropha curcas] |
| 12 |
Hb_007657_020 |
0.0755320563 |
- |
- |
PREDICTED: PRA1 family protein H isoform X1 [Jatropha curcas] |
| 13 |
Hb_000302_310 |
0.0759008647 |
- |
- |
PREDICTED: glutaminyl-peptide cyclotransferase-like [Jatropha curcas] |
| 14 |
Hb_000126_010 |
0.0781237756 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RFK1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000120_280 |
0.0786175702 |
- |
- |
PREDICTED: protein transport protein SEC23 [Jatropha curcas] |
| 16 |
Hb_000803_270 |
0.0794283087 |
- |
- |
PREDICTED: nuclear cap-binding protein subunit 1 [Jatropha curcas] |
| 17 |
Hb_002681_090 |
0.0797960738 |
- |
- |
PREDICTED: golgin candidate 6 isoform X1 [Jatropha curcas] |
| 18 |
Hb_016777_040 |
0.0804842494 |
- |
- |
PREDICTED: DNA polymerase eta-like [Jatropha curcas] |
| 19 |
Hb_005618_050 |
0.0809635526 |
- |
- |
PREDICTED: charged multivesicular body protein 7 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000005_220 |
0.0813920813 |
- |
- |
DNA ligase 1, partial [Glycine soja] |