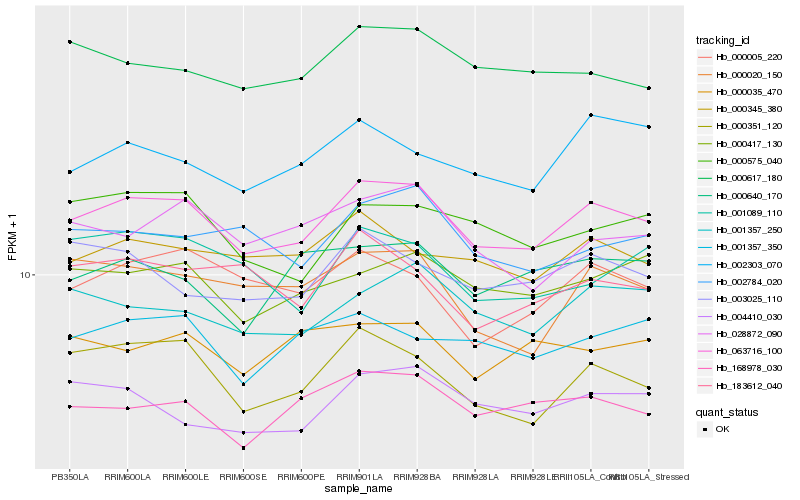
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_063716_100 |
0.0 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 56-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_000351_120 |
0.0624404653 |
- |
- |
hypothetical protein POPTR_0013s10290g [Populus trichocarpa] |
| 3 |
Hb_000417_130 |
0.0639687245 |
- |
- |
PREDICTED: SAP30-binding protein isoform X2 [Jatropha curcas] |
| 4 |
Hb_001357_250 |
0.0643740989 |
- |
- |
PREDICTED: UPF0505 protein C16orf62 homolog [Jatropha curcas] |
| 5 |
Hb_000345_380 |
0.0647887695 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 7, peroxisomal [Jatropha curcas] |
| 6 |
Hb_168978_030 |
0.0661843584 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 8 [Jatropha curcas] |
| 7 |
Hb_000617_180 |
0.0679273706 |
- |
- |
hypothetical protein B456_013G125900 [Gossypium raimondii] |
| 8 |
Hb_028872_090 |
0.0684965381 |
- |
- |
PREDICTED: urease [Jatropha curcas] |
| 9 |
Hb_000005_220 |
0.068623909 |
- |
- |
DNA ligase 1, partial [Glycine soja] |
| 10 |
Hb_000020_150 |
0.0694399668 |
- |
- |
PREDICTED: uncharacterized protein LOC105636325 [Jatropha curcas] |
| 11 |
Hb_000575_040 |
0.0694920378 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SC35 [Jatropha curcas] |
| 12 |
Hb_002784_020 |
0.0695253649 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 3 [Jatropha curcas] |
| 13 |
Hb_001357_350 |
0.0704809435 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002303_070 |
0.0706932854 |
- |
- |
PREDICTED: uncharacterized RNA-binding protein C1827.05c [Jatropha curcas] |
| 15 |
Hb_003025_110 |
0.0715721474 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_004410_030 |
0.0728144892 |
- |
- |
PREDICTED: protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS 6, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_183612_040 |
0.0736371232 |
- |
- |
PREDICTED: uncharacterized protein LOC105632370 [Jatropha curcas] |
| 18 |
Hb_000640_170 |
0.0745996296 |
- |
- |
PREDICTED: uncharacterized protein LOC105642632 isoform X2 [Jatropha curcas] |
| 19 |
Hb_001089_110 |
0.0751596321 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000035_470 |
0.0755691492 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |