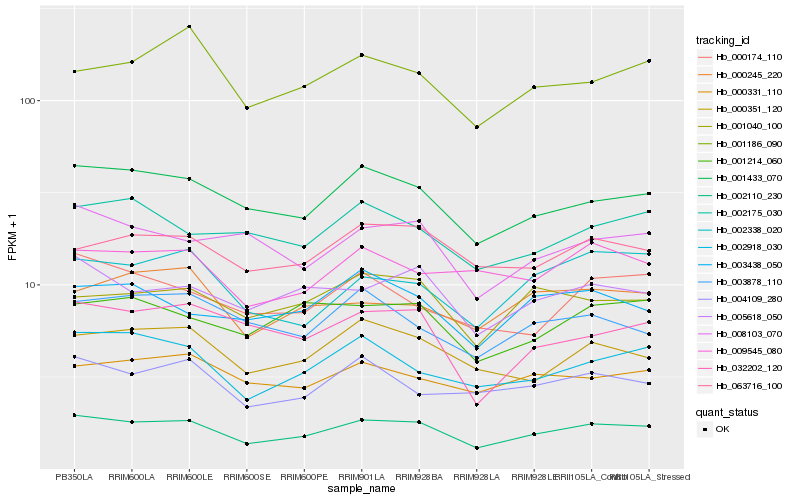
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002110_230 |
0.0 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 2 |
Hb_001433_070 |
0.0600773335 |
- |
- |
PREDICTED: splicing regulatory glutamine/lysine-rich protein 1 [Jatropha curcas] |
| 3 |
Hb_005618_050 |
0.0759928385 |
- |
- |
PREDICTED: charged multivesicular body protein 7 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000351_120 |
0.078865142 |
- |
- |
hypothetical protein POPTR_0013s10290g [Populus trichocarpa] |
| 5 |
Hb_002918_030 |
0.0808866476 |
- |
- |
PREDICTED: dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase isoform X1 [Jatropha curcas] |
| 6 |
Hb_063716_100 |
0.0838997499 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 56-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_001040_100 |
0.0847139863 |
- |
- |
PREDICTED: general transcription factor IIH subunit 4 [Jatropha curcas] |
| 8 |
Hb_001214_060 |
0.0848948891 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC8 [Jatropha curcas] |
| 9 |
Hb_000331_110 |
0.0859899894 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 10 |
Hb_003438_050 |
0.0871793694 |
- |
- |
PREDICTED: rab3 GTPase-activating protein catalytic subunit isoform X2 [Jatropha curcas] |
| 11 |
Hb_008103_070 |
0.087393219 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |
| 12 |
Hb_001186_090 |
0.0876927713 |
- |
- |
PREDICTED: nascent polypeptide-associated complex subunit alpha-like protein 2 [Jatropha curcas] |
| 13 |
Hb_004109_280 |
0.0881215111 |
- |
- |
PREDICTED: enoyl-[acyl-carrier-protein] reductase [NADH], chloroplastic-like [Populus euphratica] |
| 14 |
Hb_002175_030 |
0.0891567678 |
- |
- |
PREDICTED: snRNA-activating protein complex subunit isoform X2 [Jatropha curcas] |
| 15 |
Hb_002338_020 |
0.0892593455 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR34A [Jatropha curcas] |
| 16 |
Hb_009545_080 |
0.0904361502 |
- |
- |
hypothetical protein POPTR_0008s15400g [Populus trichocarpa] |
| 17 |
Hb_032202_120 |
0.090716299 |
- |
- |
PREDICTED: uncharacterized protein LOC105643059 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000174_110 |
0.0913874621 |
- |
- |
PREDICTED: INO80 complex subunit D-like [Jatropha curcas] |
| 19 |
Hb_000245_220 |
0.0923106797 |
- |
- |
PREDICTED: uncharacterized protein LOC105635566 [Jatropha curcas] |
| 20 |
Hb_003878_110 |
0.0923629612 |
- |
- |
PREDICTED: uncharacterized protein LOC105637024 [Jatropha curcas] |